Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 26 December 2022

A large language model for electronic health records
- Xi Yang 1 , 2 ,
- Aokun Chen 1 , 2 ,
- Nima PourNejatian 3 ,
- Hoo Chang Shin 3 ,
- Kaleb E. Smith 3 ,
- Christopher Parisien 3 ,
- Colin Compas 3 ,
- Cheryl Martin 3 ,
- Anthony B. Costa 3 ,
- Mona G. Flores ORCID: orcid.org/0000-0002-7362-3044 3 ,
- Ying Zhang ORCID: orcid.org/0000-0003-4210-2104 4 ,
- Tanja Magoc 5 ,
- Christopher A. Harle 1 , 5 ,
- Gloria Lipori 5 , 6 ,
- Duane A. Mitchell 6 ,
- William R. Hogan ORCID: orcid.org/0000-0002-9881-1017 1 ,
- Elizabeth A. Shenkman ORCID: orcid.org/0000-0003-4903-1804 1 ,
- Jiang Bian ORCID: orcid.org/0000-0002-2238-5429 1 , 2 &
- Yonghui Wu ORCID: orcid.org/0000-0002-6780-6135 1 , 2
npj Digital Medicine volume 5 , Article number: 194 ( 2022 ) Cite this article
86k Accesses
202 Citations
123 Altmetric
Metrics details
- Health care
- Medical research
There is an increasing interest in developing artificial intelligence (AI) systems to process and interpret electronic health records (EHRs). Natural language processing (NLP) powered by pretrained language models is the key technology for medical AI systems utilizing clinical narratives. However, there are few clinical language models, the largest of which trained in the clinical domain is comparatively small at 110 million parameters (compared with billions of parameters in the general domain). It is not clear how large clinical language models with billions of parameters can help medical AI systems utilize unstructured EHRs. In this study, we develop from scratch a large clinical language model—GatorTron—using >90 billion words of text (including >82 billion words of de-identified clinical text) and systematically evaluate it on five clinical NLP tasks including clinical concept extraction, medical relation extraction, semantic textual similarity, natural language inference (NLI), and medical question answering (MQA). We examine how (1) scaling up the number of parameters and (2) scaling up the size of the training data could benefit these NLP tasks. GatorTron models scale up the clinical language model from 110 million to 8.9 billion parameters and improve five clinical NLP tasks (e.g., 9.6% and 9.5% improvement in accuracy for NLI and MQA), which can be applied to medical AI systems to improve healthcare delivery. The GatorTron models are publicly available at: https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/models/gatortron_og .
Similar content being viewed by others

Generation and evaluation of artificial mental health records for Natural Language Processing
The shaky foundations of large language models and foundation models for electronic health records
Health system-scale language models are all-purpose prediction engines
Introduction.
There is an increasing interest in developing artificial intelligence (AI) systems to improve healthcare delivery and health outcomes using electronic health records (EHRs). A critical step is to extract and capture patients’ characteristics from longitudinal EHRs. The more information we have about the patients, the better the medical AI systems that we can develop. In recent decades, hospitals and medical practices in the United States (US) have rapidly adopted EHR systems 1 , 2 , resulting in massive stores of electronic patient data, including structured (e.g., disease codes, medication codes) and unstructured (i.e., clinical narratives such as progress notes). Even though using discrete data fields in clinical documentation has many potential advantages and structured data entry fields are increasingly added into the EHR systems, having clinicians use them remains a barrier, due to the added documentation burden 3 . Physicians and other healthcare providers widely use clinical narratives as a more convenient way to document patient information ranging from family medical histories to social determinants of health 4 . There is an increasing number of medical AI systems exploring the rich, more fine-grained patient information captured in clinical narratives to improve diagnostic and prognostic models 5 , 6 . Nevertheless, free-text narratives cannot be easily used in computational models that usually require structured data. Researchers have increasingly turned to natural language processing (NLP) as the key technology to enable medical AI systems to understand clinical language used in healthcare 7 .
Today, most NLP solutions are based on deep learning models 8 implemented using neural network architectures—a fast-developing sub-domain of machine learning. Convolutional neural networks 9 (CNN) and recurrent neural networks 10 (RNN) have been applied to NLP in the early stage of deep learning. More recently, the transformer architectures 11 (e.g., Bidirectional Encoder Representations from Transformers [BERT]) implemented with a self-attention mechanism 12 have become state-of-the-art, achieving the best performance on many NLP benchmarks 13 , 14 , 15 , 16 . In the general domain, the transformer-based NLP models have achieved state-of-the-art performance for name entity recognition 17 , 18 , 19 , relation extraction 20 , 21 , 22 , 23 , 24 , sentence similarity 25 , 26 , 27 , natural language inference 27 , 28 , 29 , 30 , and question answering 27 , 28 , 31 , 32 . Typically, transformers are trained in two stages: language model pretraining (i.e., learning using a self-supervised training objective on a large corpus of unlabeled text) and fine-tuning (i.e., applying the learned language models solving specific tasks with labeled training data). One pretrained language model can be applied to solve many NLP tasks through fine-tuning, which is known as transfer learning—a strategy to learn knowledge from one task and apply it in another task 33 . Human language has a very large sample space—the possible combinations of words, sentences, and their meaning and syntax are innumerable. Recent studies show that large transformer models trained using massive text data are remarkably better than previous NLP models in terms of emergence and homogenization 33 .
The promise of transformer models has led to further interest in exploring large-size (e.g., >billions of parameters) transformer models. The Generative Pretrained Transformer 3 (GPT-3) model 34 , which has 175 billion parameters and was trained using >400 billion words of text demonstrated superior performance. In the biomedical domain, researchers developed BioBERT 11 (with 110 million parameters) and PubMedBERT 35 (110 million parameters) transformer models using biomedical literature from PubMed. NVIDIA developed BioMegatron models in the biomedical domain with different sizes from 345 million to 1.2 billion parameters 36 using a more expansive set of PubMed-derived free text. However, few studies have explored scaling transformer models in the clinical domain due to the sensitive nature of clinical narratives that contain Protected Health Information (PHI) and the significant computing power required to increase the size of these models. To date, the largest transformer model using clinical narratives is ClinicalBERT 37 . ClinicalBERT has 110 million parameters and was trained using 0.5 billion words from the publicly available Medical Information Mart for Intensive Care III 38 (MIMIC-III) dataset. By developing not only larger models, but models that use clinical narratives, NLP may perform better to improve healthcare delivery and patient outcomes.
In this study, we develop a large clinical language model, GatorTron, using >90 billion words of text from the de-identified clinical notes of University of Florida (UF) Health, PubMed articles, and Wikipedia. We train GatorTron from scratch and empirically evaluate how scaling up the number of parameters benefit the performance of downstream NLP tasks. More specifically, we examine GatorTron models with varying number of parameters including (1) a base model with 345 million parameters, (2) a medium model with 3.9 billion parameters, and (3) a large model with 8.9 billion parameters. We also examine how scaling up data size benefit downstream tasks by comparing the GatorTron-base model trained from the full corpus with another GatorTron-base model trained using a random sample of 1/4 of the corpus. We compare GatorTron with existing transformer models trained using biomedical literature and clinical narratives using five clinical NLP tasks including clinical concept extraction (or named entity recognition [NER]), medical relation extraction (MRE), semantic textual similarity (STS), natural language inference (NLI), and medical question answering (MQA). GatorTron models outperform previous transformer models from the biomedical and clinical domain on five clinical NLP tasks. This study scales up transformer models in the clinical domain from 110 million to 8.9 billion parameters and demonstrates the benefit of large transformer models.
A total number of 290,482,002 clinical notes from 2,476,628 patients were extracted from the UF Health Integrated Data Repository (IDR), the enterprise data warehouse of the UF Health system. These notes were created from 2011–2021 from over 126 clinical departments and ~50 million encounters covering healthcare settings including but not limited to inpatient, outpatient, and emergency department visits. After preprocessing and de-identification, the corpus included >82 billion medical words. Figure 1 summarizes the distribution of patient by age, gender, race, and ethnicity as well as the distribution of notes by clinical department (top 5) and note type (top 5). The detailed number of patients by each category, a full list of clinical departments and the corresponding proportion of notes, and a full list of note types were provided in Supplementary Table 1 , Supplementary Table 2 , and Supplementary Table 3 .
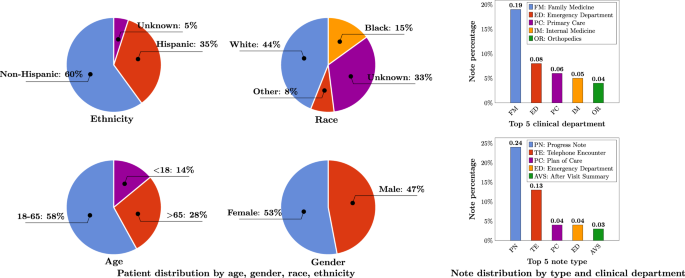
Ages were calculated as of September 2022.
Training GatorTron-large model required ~6 days on 992 A100 80 G GPUs from 124 NVIDIA DGX notes using the NVIDIA SuperPOD reference cluster architecture. Figure 2 shows the training validation loss for all three sizes of GatorTron models. The GatorTron-base model converged in 10 epochs, whereas the medium and large models converged in 7 epochs, which is consistent with prior observations on the faster per sample convergence of larger transformer models.
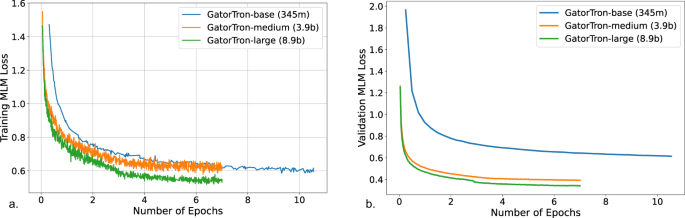
a Training loss. b Validation loss. MLM masked language modeling.
Table 1 and Table 2 compare GatorTron models with two existing biomedical transformer models (BioBERT and BioMegatron) and one clinical transformer model (Clinical BERT) on five clinical NLP tasks.
Scale up the size of training data and the number of parameters
Compared with GatorTron-base trained using a random sample of 1/4 of the corpus, the GatorTron-base model trained using the full corpus achieved improved performance for four tasks except for a sub-task in MQA (on F1 score of medication-related questions). By scaling up the number of parameters from 345 million to 8.9 billion, GatorTron-large demonstrated remarkable improvements for all five tasks, suggesting that GatorTron models scale for canonical clinical downstream tasks and that we are not yet at the limit.
Recognize clinical concepts and medical relations
Clinical concept extraction is to identify the concepts with important clinical meanings and classify their semantic categories (e.g., diseases, medications). As shown in Table 1 , all three GatorTron models outperformed existing biomedical and clinical transformer models in recognizing various types of clinical concepts on the three benchmark datasets (i.e., 2010 i2b2 39 and 2012 i2b2 40 : problem, treatments, lab tests; 2018 n2c2 41 : drug, adverse events, and drug-related attributes). The GatorTron-large model outperformed the other two smaller GatorTron models and achieved the best F1 scores of 0.8996, 0.8091, and 0.9000, respectively. For medical relation extraction—a task to identify medical relations between two clinical concepts—the GatorTron-large model also achieved the best F1 score of 0.9627 for identifying drug-cause-adverse event relations outperforming existing biomedical and clinical transformers and the other two smaller GatorTron models. We consistently observed performance improvement when scaling up the size of the GatorTron model.
Assess semantic textual similarity
The task of measuring semantic similarity is to determine the extent to which two sentences are similar in terms of semantic meaning. As shown in Table 2 , all GatorTron models outperformed existing biomedical and clinical transformer models. Among the three GatorTron models, the GatorTron-medium model achieved the best Pearson correlation score of 0.8903, outperforming both GatorTron-base and GatorTron-large. Although we did not observe consistent improvement by scaling up the size of the GatorTron model, the GatorTron-large model outperformed GatorTron-base and its performance is very close to the GatorTron-medium model (0.8896 vs. 0.8903).
Natural language inference
The task of NLI is to determine whether a conclusion can be inferred from a given sentence—a sentence-level NLP task. As shown in Table 2 , all GatorTron models outperformed existing biomedical and clinical transformers, and the GatorTron-large model achieved the best accuracy of 0.9020, outperforming the BioBERT and ClinicalBERT by 9.6% and 7.5%, respectively. We observed a monotonic performance improvement by scaling up the size of the GatorTron model.
Medical question answering
MQA is a complex clinical NLP task that requires understand information from the entire document. As shown in Table 2 , all GatorTron models outperformed existing biomedical and clinical transformer models in answering medication and relation-related questions (e.g., “What lab results does patient have that are pertinent to diabetes diagnosis?”). For medication-related questions, the GatorTron-large model achieved the best exact match score of 0.3155, outperforming the BioBERT and ClinicalBERT by 6.8% and 7.5%, respectively. For relation-related questions, GatorTron-large also achieved the best exact match score of 0.9301, outperforming BioBERT and ClinicalBERT by 9.5% and 7.77%, respectively. We also observed a monotonic performance improvement by scaling up the size of the GatorTron model.
In this study, we developed a large clinical transformer model, GatorTron, using a corpus of >90 billion words from UF Health (>82 billion), Pubmed (6 billion), Wikipedia (2.5 billion), and MIMIC III (0.5 billion). We trained GatorTron with different number of parameters including 345 million, 3.9 billion, and 8.9 billion and evaluated its performance on 5 clinical NLP tasks at different linguistic levels (phrase level, sentence level, and document level) using 6 publicly available benchmark datasets. The experimental results show that GatorTron models outperformed existing biomedical and clinical transformers for all five clinical NLP tasks evaluated using six different benchmark datasets. We observed monotonic improvements by scaling up the model size of GatorTron for four of the five tasks, excluding the semantic textual similarity task. Our GatorTron model also outperformed the BioMegatron 36 , a transformer model with a similar model size developed in our previous study using >8.5 billion words from PubMed and Wikipedia (a small proportion of the >90 billion words of corpus for developing GatorTron). This study scaled up the clinical transformer models from 345 million (ClinicalBERT) to 8.9 billion parameters in the clinical domain and demonstrated remarkable performance improvements. To the best of our knowledge, GatorTron-large is the largest transformer model in the clinical domain. Among the five tasks, GatorTron achieved remarkable improvements for complex NLP tasks such as natural language inference and medical question answering, but moderate improvements for easier tasks such as clinical concept extraction and medical relation extraction, indicating that large transformer models are more helpful to complex NLP tasks. These results are consistent with observations in the literature on the saturation of simpler benchmarks with large BERT architectures 18 , 32 .
GatorTron was pretrained using self-supervised masked language modeling (MLM) objective. We monitored training loss and calculated validation loss using a subset set of the clinical text (5%) to determine the appropriate stopping time. From the plots of training and validation losses in Fig. 2 , we observed that larger GatorTron models converged faster than the smaller model.
GatorTron models perform better in extracting and interpreting patient information documented in clinical narratives, which can be integrated into medical AI systems to improve healthcare delivery and patient outcomes. The rich, fine-grained patient information captured in clinical narratives is a critical resource powering medical AI systems. With better performance in information extraction (e.g., clinical concept extraction and medical relation extraction), GatorTron models can provide more accurate patient information to identify research-standard patient cohorts using computable phenotypes, support physicians making data-informed decisions by clinical decision support systems, and identify adverse events associated with drug exposures for pharmacovigilance. The observed improvements in semantic textual similarity, natural language inference, and medical question answering can be applied for deduplication of clinical text, mining medial knowledge, and developing next-generation medical AI systems that can interact with patients using human language.
We conducted error analysis and compared GatorTron with ClinicalBERT to probe the observed performance improvements. We found that the larger, domain-specific pretrained models (e.g., GatorTron) are better at modeling longer phrases and determining semantic categories. For example, GatorTron successfully identified “ a mildly dilated ascending aorta ”, where ClinicalBERT identified only “mildly dilated” as a problem; GatorTron successfully categorized “kidney protective effects” as a “TREATMENT”, which was mis-classified as “PROBLEM” by ClinicalBERT. For complex NLP tasks such as NLI and MQA, even large language models such as GatorTron still have difficulty in identifying the key pieces of information from longer paragraphs. Our future work will improve GatorTron in handling long pieces of text for complex NLP tasks.
This study demonstrates the advantages of large pretrained transformer models in the medical domain. GatorTron models can be applied to many other NLP tasks through fine-tuning. We believe that GatorTron will improve the use of clinical narratives in developing various medical AI systems for better healthcare delivery and health outcomes.
Data source
The primary data source for this study is the clinical narratives from UF Health IDR, a research data warehouse of UF Health. This study was approved by the UF Institutional Review Board (IRB202100049). We collected clinical notes from 2011–2021 from over 126 departments, ~2 million patients and 50 million encounters from inpatient, outpatient, and emergency settings. Then, we merged the UF Health clinical corpus with three additional corpora, including the MIMIC-III corpus 38 in the clinical domain with 0.5 billion words, a PubMed (combining PubMed abstracts and full-text commercial-collection) collection 36 in the biomedical domain with 6 billion words, and a Wikipedia articles dump 36 in the general domain with 2.5 billion words, to generate a corpus with >90 billion words.
Preprocessing and de-identification of text
We performed minimal preprocessing including (1) removing empty and duplicated clinical notes, unifying all text into UTF-8 encoding, and removing illegal UTF-8 strings; (2) normalizing special characters (e.g., convert ‘&’ to ‘&;’ ‘\xa0’ to ‘space’); (3) tokenization and sentence boundary detection. For clinical text from UF Health, we further applied a de-identification system 42 to remove protected health information (PHI) from clinical text. (Approved under IRB202100049) We adopted the safe-harbor method to identify 18 PHI categories defined in the Health Insurance Portability and Accountability Act (HIPAA) and replaced them with dummy strings (e.g., replace people’s names into [**NAME**]).
Study design
Figure 3 shows an overview of the study design. We seek to train a large clinical transformer model, GatorTron, using >90 billion words and examine how and whether scaling up model size improves performance on five clinical NLP tasks. We first pretrained GatorTron using the >90 billion words by optimizing a masked language model (MLM) and then applied GatorTron to five different clinical NLP tasks using a supervised fine-tuning. We adopted the BERT architecture (Fig. 4 ) implemented in Megatron-LM and explored three different settings including a base model of 345 million parameters (i.e., GatorTron-base), a medium model of 3.9 billion parameters (i.e., GatorTron-medium), and a large model of 8.9 billion parameters (i.e., GatorTron-large). Then we compared the three GatorTron models to an existing transformer model from the clinical domain, ClinicalBERT (trained with 110 million parameters) and two transformer models from the biomedical domain, including, BioBERT (345 million parameters) and BioMegatron (1.2 billion parameters). We compared the models on five clinical NLP tasks, including clinical concept extraction, relation extraction, semantic textual similarity, natural language inference, and medical question answering. We used six public benchmark datasets in the clinical domain.
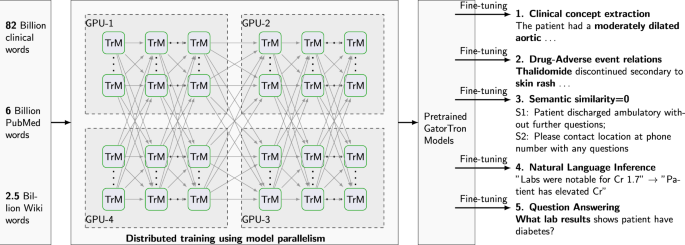
We loaded the base model and the medium model into one GPU for distributed training. We sliced the GatorTron-large model into 4 pieces and loaded model pieces to 4 GPUs for distributed training (i.e., model parallelism). TrM transformer unit.
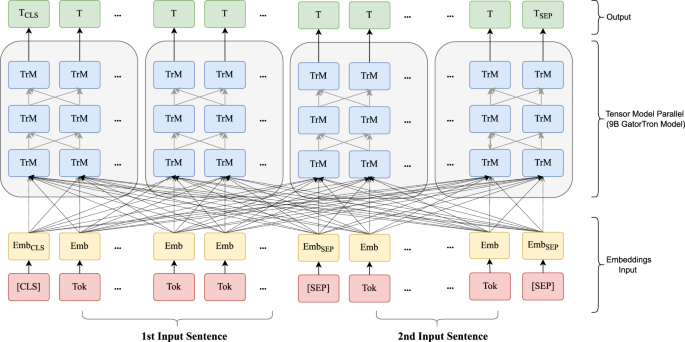
Emb embedding, Tok Token from input sentence, Trm Transformer unit. [SEP]: a token defined in BERT to indicate sentence boundaries. [CLS]: a token defined in BERT for sentence-level representation.
Training environment
We used a total number of 992 NVIDIA DGX A100 GPUs from 124 superPOD nodes at UF’s HiPerGator-AI cluster to train GatorTron models by leveraging both data-level and model-level parallelisms implemented by the Megatron-LM package 43 . We monitored the training progress by training loss and validation loss and stopped the training when there was no further improvement (i.e., the loss plot became flat).
GatorTron model configuration
We developed GatorTron models with three configurations and determined the number of layers, hidden sizes, and number of attention heads according to the guidelines for optimal depth-to-width parameter allocation proposed by Levin et al. 44 as well as our previous experience in developing BioMegatron. Table 3 provides detailed information for the three settings. The GatorTron-base model has 24 layers of transformer blocks, which is similar to the architecture of BERT-large model. For each layer, we set the number of hidden units as 1024 and attention heads as 16. The GatorTron-medium model scaled up to 3.9 billion parameters (~10 times of the base setting) and the GatorTron-large model scaled up to 8.9 billion parameters, which is similar to BioMegatron 43 (with 8.3 billion parameters).
Train GatorTron models from scratch
We pretrained a vocabulary from scratch using >90 billion words of corpus following the byte-pair-encoding algorithm 45 . We inherited the BERT-style architecture and trained GatorTron models from scratch using two self-supervised tasks, including masked language modeling (MLM) and sentence-order prediction (SOP). We followed the similar strategy in the BERT model 46 to randomly mask 15% of the input tokens with a special token (i.e., [MASK]) in the MLM. The SOP was formulated as a task to predict the order of two consecutive segments of text 28 . The input for SOP consists of two consecutive sentences from the training corpus in random orders and the training objective is to determine whether the two input sentences are in the correct order. The GatorTron-large model with 8.9 billion parameters is too large to fit one GPU, therefore, we sliced it into four pieces for distributed training using model parallelism. We pretrained the GatorTron-base and medium model without model slicing. The default loss function defined in BERT model 46 was used. Figure 4 shows the distributed training of GatorTron-large model using model parallelism. (See https://github.com/NVIDIA/Megatron-LM for more details)
Existing transformer models for comparison
BioBERT 11 : The BioBERT model was developed by further training the original BERT-large model (345 million parameters, 24 layers, 1024 hidden units, and 16 attention heads) using biomedical literature from PubMed Abstracts (4.5 billion words) and PMC Full-text articles (13.5 billion words). In this study, we used version 1.1.
ClinicalBERT 37 : The ClinicalBERT model was developed by further training the BioBERT (base version; 110 million parameters with 12 layers, 768 hidden units, and 12 attention heads) using clinical text from the MIMIC-III 38 corpus.
BioMegatron 36 : The BioMegatron models adopted the BERT architecture with a different number of parameters from 345 million to 1.2 billion. Different from BioBERT and ClinicalBERT, the BioMegatron was trained from scratch without leveraging the original BERT model.
Fine-tune GatorTron for five clinical NLP tasks, evaluation matrices, and benchmark datasets
We fine-tuned pretrained GatorTron models for five different clinical NLP tasks using experts’ annotations from six public benchmark datasets. Specifically, we first generated distributed representation from the inputs of a specific task, then added additional output layers (classification or regression) to generate target outputs. We used cross-entropy (CE) loss for classification tasks and mean square error loss for regression tasks. For a classification task with N categories, let C i be the score generated by a transformer model for category i , the probability Pi of a given sample be classified to category i was calculated as:
Let t i be the ground truth category, the cross-entropy loss L CE is defined as:
Fine-tune GatorTron for clinical concept extraction
This is a task to recognize phrases with important clinical meanings (e.g., medications, treatments, adverse drug events). The task is to determine the boundaries of a concept and classify it into predefined semantic categories. Early systems for clinical concept extract are often rule-based, yet, most recent systems are based on machine learning models such as conditional random fields (CRFs) 47 , 48 , convolutional neural networks (CNN) 9 , 49 , and recurrent neural networks (RNN) implemented with long-short-term memory strategy (LSTM) 10 , 50 . Current state-of-the-art models are based on transformers such as the ClinicalBERT. We approached clinical concept extraction as a sequence labeling problem and adopted ‘BIO’ labeling schema, where ‘B-’ and ‘I-’ are prefixes indicating words at the beginning and inside of a concept, and ‘O’ stands for words located outside of any concepts of interest. Using this definition, we approached the task as a classification problem—for each word in a sentence, predict a label in [‘B’, ‘I’, ‘O’]. When there are multiple categories of concepts, a suffix was attached to ‘BIO’ for discrimination (e.g., ‘B-drug’, ‘I-drug’). Based on the representation generated by pretrained GatorTron models, we added a classification layer (a linear layer with softmax activation) to calculate a probability score for each ‘BIO’ category. The cross-entropy loss was used for fine-tuning. We trained a unified classifier to extract all concepts for datasets without overlapped concepts. For datasets with overlapped concepts, we trained individual models to recognize each category of concept separately following our previous strategy 51 . We used three benchmark datasets developed by the 2010 i2b2 challenge 39 , 2012 i2b2 challenge 40 , and 2018 n2c2 challenge 41 to evaluate GatorTron models focusing on identifying important medical concepts (e.g., medications, adverse drug events, treatments) from clinical text. We used precision, recall, and F1 score for evaluation.
Fine-tune GatorTron for medical relation extraction
MRE is to establish medical-related relations (e.g., induce relation) among clinical concepts (e.g., drugs, adverse events). MRE is usually approached as a classification problem—identify pairs of concepts with valid relations and classify the relation type. Various machine learning-based classifiers such as support vector machines (SVMs), random forests (RF), and gradient boosting trees (GBT) 41 have been applied. With the emergence of deep learning models, researchers have explored the long-short-term memory (LSTM) architecture for RE in both general and clinical domains 52 , 53 . Most recently, several studies adopted the BERT architecture and demonstrated superior performance for MRE on various datasets 54 , 55 , 56 , 57 , 58 , 59 . We approached MRE as a classification task. First, candidate concept pairs were generated using heuristic rules developed in our previous study 41 . Then, we identified two sentences where the two concepts in a pair were located. We introduced two sets of entity markers (i.e., [S1], [E1] and [S2], [E2]) to indicate the two concepts. If the two concepts were in the same sentence, the two input sentences will be the same but labeled with different markers (e.g., [S1] and [E1] were used in the first sentence; [S2] and [E2] were used in the second sentence). To determine the relation type, we concatenated the representations of the model special [CLS] token and all four entity markers and added a classification layer (a linear layer with softmax activation) for classification. Similarly, the cross-entropy loss was used to fine-tune GatorTron. We used the dataset developed by the 2018 n2c2 challenge 41 with a focus on relations between medications and adverse drug events. The precision, recall, and F1 score were used for evaluation.
Fine-tune GatorTron for semantic textual similarity
The STS task is to quantitatively assess the semantic similarity between two text snippets (e.g., sentences), which is usually approached as a regression task where a real-value score was used to quantify the similarity between two text snippets. In the general domain, the STS benchmark (STS-B) dataset curated by the Semantic Evaluation (SemEval) challenges between 2012 and 2017 60 is widely used for evaluating STS systems 13 . Various machine learning methods have been examined 61 , 62 , 63 but transformer-based systems such as RoBERTa 25 , T5 27 , and ALBERT 28 are leading the state-of-the-art models for STS. In the clinical domain, the MedSTS dataset 64 that consists of over 1000 annotated sentence pairs from clinical notes at Mayo Clinic was widely used as the benchmark. MedSTS was used as the gold standard in two clinical NLP open challenges including the 2018 BioCreative/Open Health NLP (OHNLP) challenge 65 and 2019 n2c2/OHNLP ClinicalSTS shared task 66 . Similar to the general domain, pretrained transformer-based models using clinical text and biomedical literature, including ClinicalBERT and BioBERT 67 , achieved state-of-the-art performance. In this study, we formulated STS as a regression problem. We applied pretrained GatorTron models to learn the sentence-level representations of the two pieces of text and adopted a linear regression layer to calculate the similarity score. Different from classification models, we used MSE as the loss function. We used the dataset developed by the 2019 n2c2/OHNLP 66 challenge on clinical semantic textural similarity 66 . The Pearson correlation score was used for evaluation.
Fine-tune GatorTron for natural language inference
NLI is also known as recognizing textual entailment (RTE)—a directional relation between text fragments (e.g., sentences) 68 . The goal of NLI is to determine if a given hypothesis can be inferred from a given premise. In the general domain, two benchmark datasets—the MultiNLI 69 and the Stanford NLI 70 are widely used. On both datasets, pretrained transformer models achieved state-of-the-art performances 27 , 29 . There are limited resources for NLI in the clinical domain. Until recently, the MedNLI—a dataset annotated by doctors based on the medical history of patients 71 was developed as a benchmark dataset in the clinical domain. A previous study 37 showed that a pretrained clinical BERT model achieved the state-of-the-art performance and outperformed the baseline (InferSent 72 ) by ~9% accuracy. In this study, we approached NLI as a classification problem. We concatenated the hypothesis and premise as the input separated using a special token [SEP] and applied pretrained GatorTron models to generate distributed representations, which were fed into a classification layer (a linear layer with softmax activation) to calculate a probability for each of the three categories of entailment, contradiction, and neutral. The cross-entropy loss was used for fine-tuning. We evaluated the GatorTron models on NLI using the MedNLI dataset 71 and used accuracy for comparison.
Fine-Tune GatorTron for medical question answering
The MQA task is to build NLP systems that automatically answer medical questions in a natural language, which is the most complex challenge among the five tasks. Unlike other tasks focusing on phrases and sentences, MQA is a document-level task that requires information from the whole document to generate answers according to questions. In the general domain, the Stanford Question Answering Datasets (SQuAD 1.1 and 2.0) 73 , 74 have been widely used as benchmarks. Transformer-based models are state-of-the-art for both SQuAD1.1 18 and SQuAD2.0 31 . There are several MQA datasets developed in the past few years such as the MESHQA 75 , MedQuAD 76 , and emrQA 77 . In this study, we approached MQA using a machine reading comprehension (MRC) technique where the goal is to extract the most relevant responses (i.e., short text snippets or entities) from the given context according to questions. We applied a span classification algorithm to identify the start and end offsets of the answer from the context. More specifically, we packed the question and the context into a single sequence as input for GatorTron and applied two linear layers to predict the start and end position of the answer, respectively. As GatorTron models were developed using a maximum token length of 512, we limited the maximum length of questions to 64 tokens and the rest of the 446 tokens (including special tokens such as [CLS] and [SEP]) were used for the context. We truncated questions with more than 64 tokens. For contexts the had more than 446 tokens, we adopted a sliding window strategy to scan the whole document using a window size of 446 tokens and a stride size of 396 tokens, so that two consecutive windows had the same 50 tokens overlapped. We also limited the answers to a maximum length of 32 tokens. We used the emrQA dataset 77 , which is widely used as a benchmark dataset for MQA. We particularly focused on medications and relations-related questions as Yue et al. 78 found that the two subsets are more consistent. We utilized both F1 score and exact match score for evaluation.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The benchmark datasets that support the findings of this study are available from the official websites of natural language processing challenges with Data Use Agreements. More specifically: (1) i2b2 2010, 2012 datasets and n2c2 2018, 2019 datasets: https://portal.dbmi.hms.harvard.edu/projects/n2c2-nlp/ . (2) MedNLI dataset: https://physionet.org/content/mednli/1.0.0/ . (3) emrQA dataset: https://github.com/panushri25/emrQA#download-dataset . (4) MIMIC III dataset: https://physionet.org/content/mimiciii/1.4/ . (5) PubMed dataset: https://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/ . (6) Wikipedia dataset: https://dumps.wikimedia.org/enwiki/latest/enwiki-latest-pages-articles.xml.bz2 . (7) UF Health IDR clinical notes are not open to the public due to patient privacy information. The GatorTron models pretrained using >90 billion words of text is publicly available at: https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/models/gatortron_og .
Code availability
The computer codes to train GatorTron models are available from: https://github.com/NVIDIA/Megatron-LM and https://github.com/NVIDIA/NeMo . The computer codes for preprocessing of text data are available from: https://github.com/uf-hobi-informatics-lab/NLPreprocessing https://github.com/uf-hobi-informatics-lab/GatorTron .
Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals: 2008–2015. ONC Data Brief . https://www.healthit.gov/sites/default/files/briefs/2015_hospital_adoption_db_v17.pdf (2016).
Adler-Milstein, J. et al. Electronic health record adoption in US hospitals: the emergence of a digital ‘advanced use’ divide. J. Am. Med. Inform. Assoc. 24 , 1142–1148 (2017).
Article Google Scholar
Bush, R. A., Kuelbs, C. L., Ryu, J., Jian, W. & Chiang, G. J. Structured data entry in the electronic medical record: perspectives of pediatric specialty physicians and surgeons. J. Med. Syst. 41 , 1–8 (2017).
Meystre, S. M., Savova, G. K., Kipper-Schuler, K. C. & Hurdle, J. F. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb. Med. Inform. 17 , 128–144 (2008).
Liang, H. et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. 25 , 433–438 (2019).
Article CAS Google Scholar
Yang, J. et al. Assessing the prognostic significance of tumor-infiltrating lymphocytes in patients with melanoma using pathologic features identified by natural language processing. JAMA Netw. Open 4 , e2126337 (2021).
Nadkarni, P. M., Ohno-Machado, L. & Chapman, W. W. Natural language processing: an introduction. J. Am. Med. Inform. Assoc. 18 , 544–551 (2011).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521 , 436–444 (2015).
Collobert, R. et al. Natural language processing (almost) from scratch. J. Mach. Learn Res. 12 , 2493–2537 (2011).
Google Scholar
Lample, G., Ballesteros, M., Subramanian, S., Kawakami, K. & Dyer, C. Neural architectures for named entity recognition. Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies . 260–270 (2016).
Lee, J. et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 36 , 1234–1240 (2020).
CAS Google Scholar
Vaswani, A. et al. Attention is All you Need. Advances in Neural Information Processing Systems . 30 (2017).
Wang, A. et al. GLUE: A multi-task benchmark and analysis platform for natural language understanding. Proceedings of the 2018 EMNLP Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP. 353–355 (2018).
Wang, A. et al. SuperGLUE: a stickier benchmark for general-purpose language understanding systems. Advances in neural information processing systems . 32 (2019).
Qiu, X. et al. Pre-trained models for natural language processing: a survey. Science China Technological Sciences. 63 , 1872–1897 (2020).
Tay, Y., Dehghani, M., Bahri, D. & Metzler, D. Efficient transformers: a survey. ACM Computing Surveys. 55 , 1–28 (2020).
Yu, J., Bohnet, B. & Poesio, M. Named entity recognition as dependency parsing. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics . 6470–6476 (2020).
Yamada, I., Asai, A., Shindo, H., Takeda, H. & Matsumoto, Y. LUKE: deep contextualized entity representations with entity-aware self-attention. Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) . 6442–6454 (2020).
Li, X. et al. Dice loss for data-imbalanced NLP tasks. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics . 465–476 (2020).
Xu, B., Wang, Q., Lyu, Y., Zhu, Y. & Mao, Z. Entity structure within and throughout: modeling mention dependencies for document-level relation extraction. Proceedings of the AAAI Conference on Artificial Intelligence 35 , 14149–14157 (2021).
Ye, D., Lin, Y. & Sun, M. Pack together: entity and relation extraction with levitated marker. Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics . 1 , 4904–4917 (2021).
Cohen, A. D., Rosenman, S. & Goldberg, Y. Relation classification as two-way span-prediction. ArXiv arXiv:2010.04829 (2021).
Lyu, S. & Chen, H. Relation classification with entity type restriction. Findings of the Association for Computational Linguistics: ACL-IJCNLP . 390–395 (2021).
Wang, J. & Lu, W. Two are better than one: joint entity and relation extraction with table-sequence encoders. Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) . 1706–1721 (2020).
Jiang, H. et al. SMART: Robust and efficient fine-tuning for pre-trained natural language models through principled regularized optimization. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. 2177–2190 (2020).
Yang, Z. et al. XLNet: Generalized Autoregressive Pretraining for Language Understanding. Proceedings of the 33rd International Conference on Neural Information Processing Systems . 5753–5763 (2019).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21 , 1–67 (2019).
Lan, Z.-Z. et al. ALBERT: a lite BERT for self-supervised learning of language representations. ArXiv arXiv: 1909.11942 (2019).
Wang, S., Fang, H., Khabsa, M., Mao, H. & Ma, H. Entailment as Few-Shot Learner. ArXiv arXiv: 2104.14690 (2021).
Zhang, Z. et al. Semantics-aware BERT for language understanding. Proceedings of the AAAI Conference on Artificial Intelligence . 34 , 9628-963 5 (2020).
Zhang, Z., Yang, J. & Zhao, H. Retrospective reader for machine reading comprehension. Proceedings of the AAAI Conference on Artificial Intelligence . 35 , 14506-14514 (2021).
Garg, S., Vu, T. & Moschitti, A. TANDA: transfer and adapt pre-trained transformer models for answer sentence selection. Proceedings of the AAAI Conference on Artificial Intelligence. 34, 7780-7788 (2020).
Bommasani, R. et al. On the opportunities and risks of foundation models. ArXiv arXiv: 2108.07258 (2021).
Floridi, L. & Chiriatti, M. GPT-3: its nature, scope, limits, and consequences. Minds Mach 30 , 681–694 (2020).
Gu, Y. et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthc. 3 , 1–23 (2022).
Shin, H.-C. et al. BioMegatron: larger biomedical domain language model. Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) . 4700–4706 (2020).
Alsentzer, E. et al. Publicly Available Clinical BERT Embeddings. in Proc. 2nd Clinical Natural Language Processing Workshop 72–78 (2019).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3 , 160035 (2016).
Uzuner, Ö., South, B. R., Shen, S. & DuVall, S. L. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc. 18 , 552–556 (2011).
Sun, W., Rumshisky, A. & Uzuner, O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J. Am. Med. Inform. Assoc. 20 , 806–813 (2013).
Yang, X. et al. Identifying relations of medications with adverse drug events using recurrent convolutional neural networks and gradient boosting. J. Am. Med. Inform. Assoc. 27 , 65–72 (2020).
Yang, X. et al. A study of deep learning methods for de-identification of clinical notes in cross-institute settings. BMC Med. Inform. Decis. Mak. 19 , 232 (2019).
Shoeybi, M. et al. Megatron-LM: training multi-billion parameter language models using model parallelism. ArXiv arXiv:1909.08053 (2020).
Levine, Y., Wies, N., Sharir, O., Bata, H. & Shashua, A. Limits to depth efficiencies of self-attention. Advances in Neural Information Processing Systems 33 , 22640–22651 (2020).
Sennrich, R., Haddow, B. & Birch, A. Neural Machine Translation of Rare Words with Subword Units. in Proc. 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) 1715–1725 (Association for Computational Linguistics, 2016).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies . 4171–4186 (2019).
Wu, Y., Xu, J., Jiang, M., Zhang, Y. & Xu, H. A study of neural word embeddings for named entity recognition in clinical text. Amia. Annu. Symp. Proc. 2015 , 1326–1333 (2015).
Soysal, E. et al. CLAMP—a toolkit for efficiently building customized clinical natural language processing pipelines. J. Am. Med. Inform. Assoc. 25 , 331–336 (2018).
Wu, Y., Jiang, M., Lei, J. & Xu, H. Named entity recognition in chinese clinical text using deep neural network. Stud. Health Technol. Inform. 216 , 624–628 (2015).
Wu, Y. et al. Combine factual medical knowledge and distributed word representation to improve clinical named entity recognition. in AMIA Annual Symposium Proceedings vol. 2018, 1110 (American Medical Informatics Association, 2018).
Kumar, S. A survey of deep learning methods for relation extraction. ArXiv arXiv:1705.03645 (2017).
Lv, X., Guan, Y., Yang, J. & Wu, J. Clinical relation extraction with deep learning. Int. J. Hybrid. Inf. Technol. 9 , 237–248 (2016).
Wei, Q. et al. Relation extraction from clinical narratives using pre-trained language models. Amia. Annu. Symp. Proc. 2019 , 1236–1245 (2020).
Guan, H. & Devarakonda, M. Leveraging contextual information in extracting long distance relations from clinical notes. Amia. Annu. Symp. Proc. 2019 , 1051–1060 (2020).
Alimova, I. & Tutubalina, E. Multiple features for clinical relation extraction: a machine learning approach. J. Biomed. Inform. 103 , 103382 (2020).
Mahendran, D. & McInnes, B. T. Extracting adverse drug events from clinical notes. AMIA Summits on Translational Science Proceedings . 420–429 (2021).
Yang, X., Zhang, H., He, X., Bian, J. & Wu, Y. Extracting family history of patients from clinical narratives: exploring an end-to-end solution with deep learning models. JMIR Med. Inform. 8 , e22982 (2020).
Yang, X., Yu, Z., Guo, Y., Bian, J. & Wu, Y. Clinical Relation Extraction Using Transformer-based Models. ArXiv. arXiv:2107.08957 (2021).
Cer, D., Diab, M., Agirre, E., Lopez-Gazpio, I. & Specia, L. Semeval-2017 task 1: Semantic textual similarity-multilingual and cross-lingual focused evaluation. Proceedings of the 11th International Workshop on Semantic Evaluation (SemEval-2017) . 1–14 (2017).
Farouk, M. Measuring sentences similarity: a survey. ArXiv arXiv:1910.03940 (2019).
Ramaprabha, J., Das, S. & Mukerjee, P. Survey on sentence similarity evaluation using deep learning. J. Phys. Conf. Ser. 1000 , 012070 (2018).
Gomaa, W. H. & Fahmy, A. A survey of text similarity approaches. International journal of Computer Applications 68 , 13–18 (2013).
Wang, Y. et al. MedSTS: a resource for clinical semantic textual similarity. Lang. Resour. Eval. 54 , 57–72 (2020).
Rastegar-Mojarad, M. et al. BioCreative/OHNLP Challenge 2018. in Proc. 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics 575–575 (ACM, 2018).
Wang, Y. et al. Overview of the 2019 n2c2/OHNLP track on clinical semantic textual similarity. JMIR Med. Inform. 8 , e23375 (2020).
Mahajan, D. et al. Identification of semantically similar sentences in clinical notes: iterative intermediate training using multi-task learning. JMIR Med. Inform. 8 , e22508 (2020).
Dagan, I., Glickman, O. & Magnini, B. in Machine Learning Challenges. Evaluating Predictive Uncertainty, Visual Object Classification, and Recognising Tectual Entailment (eds. Quiñonero-Candela, J., Dagan, I., Magnini, B. & d’Alché-Buc, F.) 177–190 (Springer Berlin Heidelberg, 2006).
Williams, A., Nangia, N. & Bowman, S. R. A broad-coverage challenge corpus for sentence understanding through inference. Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies . 1 , 1112–1122 (2018).
Bowman, S. R., Angeli, G., Potts, C. & Manning, C. D. A large annotated corpus for learning natural language inference. Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing . 632–642 (2015).
Shivade, C. MedNLI—a natural language inference dataset for the clinical domain. PhysioNet https://doi.org/10.13026/C2RS98 (2017).
Conneau, A., Kiela, D., Schwenk, H., Barrault, L. & Bordes, A. Supervised learning of universal sentence representations from natural language inference data. Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing . 670–680 (2017).
Rajpurkar, P., Zhang, J., Lopyrev, K. & Liang, P. SQuAD: 100,000+ questions for machine comprehension of text. Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing . 2383–2392 (2016).
Rajpurkar, P., Jia, R. & Liang, P. Know what you don’t know: unanswerable questions for SQuAD. Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics 2 , 784–789 (2018).
Zhu, M., Ahuja, A., Juan, D.-C., Wei, W. & Reddy, C. K. Question Answering with Long Multiple-Span Answers. in Findings of the Association for Computational Linguistics: EMNLP 2020 3840–3849 (Association for Computational Linguistics, 2020).
Ben Abacha, A. & Demner-Fushman, D. A question-entailment approach to question answering. BMC Bioinforma 20 , 511 (2019).
Pampari, A., Raghavan, P., Liang, J. & Peng, J. emrQA: a large corpus for question answering on electronic medical records. Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing . 2357–2368 (2018).
Yue, X., Gutierrez, B. J. & Sun, H. Clinical reading comprehension: a thorough analysis of the emrQA dataset. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics . 4474–4486 (2020).
Download references
Acknowledgements
This study was partially supported by a Patient-Centered Outcomes Research Institute® (PCORI®) Award (ME-2018C3-14754), a grant from the National Cancer Institute, 1R01CA246418 R01, grants from the National Institute on Aging, NIA R56AG069880 and R21AG062884, and the Cancer Informatics and eHealth core jointly supported by the UF Health Cancer Center and the UF Clinical and Translational Science Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutions. We would like to thank the UF Research Computing team, led by Dr. Erik Deumens, for providing computing power through UF HiPerGator-AI cluster.
Author information
Authors and affiliations.
Department of Health Outcomes and Biomedical Informatics, College of Medicine, University of Florida, Gainesville, FL, USA
Xi Yang, Aokun Chen, Christopher A. Harle, William R. Hogan, Elizabeth A. Shenkman, Jiang Bian & Yonghui Wu
Cancer Informatics and eHealth core, University of Florida Health Cancer Center, Gainesville, FL, USA
Xi Yang, Aokun Chen, Jiang Bian & Yonghui Wu
NVIDIA, Santa Clara, CA, USA
Nima PourNejatian, Hoo Chang Shin, Kaleb E. Smith, Christopher Parisien, Colin Compas, Cheryl Martin, Anthony B. Costa & Mona G. Flores
Research Computing, University of Florida, Gainesville, FL, USA
Integrated Data Repository Research Services, University of Florida, Gainesville, FL, USA
Tanja Magoc, Christopher A. Harle & Gloria Lipori
Lillian S. Wells Department of Neurosurgery, UF Clinical and Translational Science Institute, University of Florida, Gainesville, FL, USA
Gloria Lipori & Duane A. Mitchell
You can also search for this author in PubMed Google Scholar
Contributions
Y.W., J.B., M.G.F., N.P., and X.Y. were responsible for the overall design, development, and evaluation of this study. X.Y. and A.C. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Y.W., X.Y., J.B., and W.H. did the bulk of the writing, E.A.S., D.A.M., T.M., C.A.H., A.B.C., and G.L. also contributed to writing and editing of this manuscript. All authors reviewed the manuscript critically for scientific content, and all authors gave final approval of the manuscript for publication.
Corresponding author
Correspondence to Yonghui Wu .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental material, reporting summary, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Yang, X., Chen, A., PourNejatian, N. et al. A large language model for electronic health records. npj Digit. Med. 5 , 194 (2022). https://doi.org/10.1038/s41746-022-00742-2
Download citation
Received : 21 June 2022
Accepted : 13 December 2022
Published : 26 December 2022
DOI : https://doi.org/10.1038/s41746-022-00742-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Assessing the research landscape and clinical utility of large language models: a scoping review.
- Ye-Jean Park
- Abhinav Pillai
- Christopher Naugler
BMC Medical Informatics and Decision Making (2024)
Generative AI in healthcare: an implementation science informed translational path on application, integration and governance
- Sandeep Reddy
Implementation Science (2024)
Research ethics and artificial intelligence for global health: perspectives from the global forum on bioethics in research
- Effy Vayena
BMC Medical Ethics (2024)
MetaTron: advancing biomedical annotation empowering relation annotation and collaboration
- Ornella Irrera
- Stefano Marchesin
- Gianmaria Silvello
BMC Bioinformatics (2024)
Transformer models in biomedicine
- Sumit Madan
- Manuel Lentzen
- Holger Fröhlich
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Open access
- Published: 27 October 2021
A narrative review on the validity of electronic health record-based research in epidemiology
- Milena A. Gianfrancesco 1 &
- Neal D. Goldstein ORCID: orcid.org/0000-0002-9597-5251 2
BMC Medical Research Methodology volume 21 , Article number: 234 ( 2021 ) Cite this article
12k Accesses
58 Citations
5 Altmetric
Metrics details
Electronic health records (EHRs) are widely used in epidemiological research, but the validity of the results is dependent upon the assumptions made about the healthcare system, the patient, and the provider. In this review, we identify four overarching challenges in using EHR-based data for epidemiological analysis, with a particular emphasis on threats to validity. These challenges include representativeness of the EHR to a target population, the availability and interpretability of clinical and non-clinical data, and missing data at both the variable and observation levels. Each challenge reveals layers of assumptions that the epidemiologist is required to make, from the point of patient entry into the healthcare system, to the provider documenting the results of the clinical exam and follow-up of the patient longitudinally; all with the potential to bias the results of analysis of these data. Understanding the extent of as well as remediating potential biases requires a variety of methodological approaches, from traditional sensitivity analyses and validation studies, to newer techniques such as natural language processing. Beyond methods to address these challenges, it will remain crucial for epidemiologists to engage with clinicians and informaticians at their institutions to ensure data quality and accessibility by forming multidisciplinary teams around specific research projects.
Peer Review reports
The proliferation of electronic health records (EHRs) spurred on by federal government incentives over the past few decades has resulted in greater than an 80% adoption-rate at hospitals [ 1 ] and close to 90% in office-based practices [ 2 ] in the United States. A natural consequence of the availability of electronic health data is the conduct of research with these data, both observational and experimental [ 3 ], due to lower overhead costs and lower burden of study recruitment [ 4 ]. Indeed, a search on PubMed for publications indexed by the MeSH term “electronic health records” reveals an exponential growth in biomedical literature, especially over the last 10 years with an excess of 50,000 publications.
An emerging literature is beginning to recognize the many challenges that still lay ahead in using EHR data for epidemiological investigations. Researchers in Europe identified 13 potential sources of “bias” (bias was defined as a contamination of the data) in EHR-based data covering almost every aspect of care delivery, from selective entrance into the healthcare system, to variation in care and documentation practices, to identification and extraction of the right data for analysis [ 5 ]. Many of the identified contaminants are directly relevant to traditional epidemiological threats to validity [ 4 ]. Data quality has consistently been invoked as a central challenge in EHRs. From a qualitative perspective, healthcare workers have described challenges in the healthcare environment (e.g., heavy workload), imperfect clinical documentation practices, and concerns over data extraction and reporting tools, all of which would impact the quality of data in the EHR [ 6 ]. From a quantitative perspective, researchers have noted limited sensitivity of diagnostic codes in the EHR when relying on discrete codings, noting that upon a manual chart review free text fields often capture the missed information, motivating such techniques as natural language processing (NLP) [ 7 ]. A systematic review of EHR-based studies also identified data quality as an overarching barrier to the use of EHRs in managing the health of the community, i.e. “population health” [ 8 ]. Encouragingly this same review also identified more facilitators than barriers to the use of EHRs in public health, suggesting that opportunities outweigh the challenges. Shortreed et al. further explored these opportunities discussing how EHRs can enhance pragmatic trials, bring additional sophistication to observational studies, aid in predictive modeling, and be linked together to create more comprehensive views of patients’ health [ 9 ]. Yet, as Shortreed and others have noted, significant challenges still remain.
It is our intention with this narrative review to discuss some of these challenges in further detail. In particular, we focus on specific epidemiological threats to validity -- internal and external -- and how EHR-based epidemiological research in particular can exacerbate some of these threats. We note that while there is some overlap in the challenges we discuss with traditional paper-based medical record research that has occurred for decades, the scale and scope of an EHR-based study is often well beyond what was traditionally possible in the manual chart review era and our applied examples attempt to reflect this. We also describe existing and emerging approaches for remediating these potential biases as they arise. A summary of these challenges may be found in Table 1 . Our review is grounded in the healthcare system in the United States, although we expect many of the issues we describe to be applicable regardless of locale; where necessary, we have flagged our comments as specific to the U.S.
Challenge #1: Representativeness
The selection process for how patients are captured in the EHR is complex and a function of geographic, social, demographic, and economic determinants [ 10 ]. This can be termed the catchment of the EHR. For a patient record to appear in the EHR the patient must have been registered in the system, typically to capture their demographic and billing information, and upon a clinical visit, their health details. While this process is not new to clinical epidemiology, what tends to separate EHR-based records from traditional paper-based records is the scale and scope of the data. Patient data may be available for longer periods of time longitudinally, as well as have data corresponding to interactions with multiple, potentially disparate, healthcare systems [ 11 ]. Given the consolidation of healthcare [ 12 ] and aggregated views of multiple EHRs through health information networks or exchanges [ 11 ] the ability to have a complete view of the patients’ total health is increasing. Importantly, the epidemiologist must ascertain whether the population captured within the EHR or EHR-derived data is representative of the population targeted for inference. This is particularly true under the paradigm of population health and inferring the health status of a community from EHR-based records [ 13 ]. For example, a study of Clostridium difficile infection at an urban safety net hospital in Philadelphia, Pennsylvania demonstrated notable differences in risk factors in the hospital’s EHR compared to national surveillance data, suggesting how catchment can influence epidemiologic measures [ 14 ]. Even health-related data captured through health information exchanges may be incomplete [ 15 ].
Several hypothetical study settings can further help the epidemiologist appreciate the relationship between representativeness and validity in EHR research. In the first hypothetical, an EHR-based study is conducted from a single-location federally qualified health center, and in the second hypothetical, an EHR-based study is conducted from a large academic health system. Suppose both studies occur in the same geographic area. It is reasonable to believe the patient populations captured in both EHRs will be quite different and the catchment process could lead to divergent estimates of disease or risk factor prevalence. The large academic health system may be less likely to capture primary care visits, as specialty care may drive the preponderance of patient encounters. However, this is not a bias per se : if the target of inference from these two hypothetical EHR-based studies is the local community, then selection bias becomes a distinct possibility. The epidemiologist must also consider the potential for generalizability and transportability -- two facets of external validity that respectively relate to the extrapolation of study findings to the source population or a different population altogether -- if there are unmeasured effect modifiers, treatment interference, or compound treatments in the community targeted for inference [ 16 ].
There are several approaches for ascertaining representativeness of EHR-based data. Comparing the EHR-derived sample to Census estimates of demography is straightforward but has several important limitations. First, as previously described, the catchment process may be driven by discordant geographical areas, especially for specialty care settings. Second and third, the EHR may have limited or inaccurate information on socioeconomic status, race, and ethnicity that one may wish to compare [ 17 , 18 ], and conversely the Census has limited estimates of health, chiefly disability, fertility, and insurance and payments [ 19 ]. If selection bias is suspected as a result of missing visits in a longitudinal study [ 20 ] or the catchment process in a cross-sectional study [ 21 ], using inverse probability weighting may remediate its influence. Comparing the weighted estimates to the original, non-weighted estimates provides insight into differences in the study participants. In the population health paradigm whereby the EHR is used as a surveillance tool to identify community health disparities [ 13 ], one also needs to be concerned about representativeness. There are emerging approaches for producing such small area community estimates from large observational datasets [ 22 , 23 ]. Conceivably, these approaches may also be useful for identifying issues of representativeness, for example by comparing stratified estimates across sociodemographic or other factors that may relate to catchment. Approaches for issues concerning representativeness specifically as it applies to external validity may be found in these references [ 24 , 25 ].
Challenge #2: Data availability and interpretation
Sub-challenge #2.1: billing versus clinical versus epidemiological needs.
There is an inherent tension in the use of EHR-based data for research purposes: the EHR was never originally designed for research. In the U.S., the Health Information Technology for Economic and Clinical Health Act, which promoted EHRs as a platform for comparative effectiveness research, was an attempt to address this deficiency [ 26 ]. A brief history of the evolution of the modern EHR reveals a technology that was optimized for capturing health details relevant for billing, scheduling, and clinical record keeping [ 27 ]. As such, the availability of data for fundamental markers of upstream health that are important for identifying inequities, such as socioeconomic status, race, ethnicity, and other social determinants of health (SDOH), may be insufficiently captured in the EHR [ 17 , 18 ]. Similarly, behavioral risk factors, such as being a sexual minority person, have historically been insufficiently recorded as discrete variables. It is only recently that such data are beginning to be captured in the EHR [ 28 , 29 ], or techniques such as NLP have made it possible to extract these details when stored in free text notes (described further in “ Unstructured data: clinical notes and reports ” section).
As an example, assessing clinical morbidities in the EHR may be done on the basis of extracting appropriate International Classification of Diseases (ICD) codes, used for billing and reimbursement in the U.S. These codes are known to have low sensitivity despite high specificity for accurate diagnostic status [ 30 , 31 ]. Expressed as predictive values, which depend upon prevalence, presence of a diagnostic code is a likely indicator of a disease state, whereas absence of a diagnostic code is a less reliable indicator of the absence of that morbidity. There may further be variation by clinical domain in that ICD codes may exist but not be used in some specialties [ 32 ], variation by coding vocabulary such as the use of SNOMED for clinical documentation versus ICD for billing necessitating an ontology mapper [ 33 ], and variation by the use of “rule-out” diagnostic codes resulting in false-positive diagnoses [ 34 , 35 , 36 ]. Relatedly is the notion of upcoding, or the billing of tests, procedures, or diagnoses to receive inflated reimbursement, which, although posited to be problematic in EHRs [ 37 ] in at least one study, has not been shown to have occurred [ 38 ]. In the U.S., the billing and reimbursement model, such as fee-for-service versus managed care, may result in varying diagnostic code sensitivities and specificities, especially if upcoding is occurring [ 39 ]. In short, there is potential for misclassification of key health data in the EHR.
Misclassification can potentially be addressed through a validation study (resources permitting) or application of quantitative bias analysis, and there is a rich literature regarding the treatment of misclassified data in statistics and epidemiology. Readers are referred to these texts as a starting point [ 40 , 41 ]. Duda et al. and Shepherd et al. have described an innovative data audit approach applicable to secondary analysis of observational data, such as EHR-derived data, that incorporates the audit error rate directly in the regression analysis to reduce information bias [ 42 , 43 ]. Outside of methodological tricks in the face of imperfect data, researchers must proactively engage with clinical and informatics colleagues to ensure that the right data for the research interests are available and accessible.
Sub-challenge #2.2: Consistency in data and interpretation
For the epidemiologist, abstracting data from the EHR into a research-ready analytic dataset presents a host of complications surrounding data availability, consistency and interpretation. It is easy to conflate the total volume of data in the EHR with data that are usable for research, however expectations should be tempered. Weiskopf et al. have noted such challenges for the researcher: in their study, less than 50% of patient records had “complete” data for research purposes per their four definitions of completeness [ 44 ]. Decisions made about the treatment of incomplete data can induce selection bias or impact precision of estimates (see Challenges #1 , #3 , and #4 ). The COVID-19 pandemic has further demonstrated the challenge of obtaining research data from EHRs across multiple health systems [ 45 ]. On the other hand, EHRs have a key advantage of providing near real-time data as opposed to many epidemiological studies that have a specific endpoint or are retrospective in nature. Such real-time data availability was leveraged during COVID-19 to help healthcare systems manage their pandemic response [ 46 , 47 ]. Logistical and technical issues aside, healthcare and documentation practices are nuanced to their local environments. In fact, researchers have demonstrated how the same research question analyzed in distinct clinical databases can yield different results [ 48 ].
Once the data are obtained, choices regarding operationalization of variables have the potential to induce information bias. Several hypothetical examples can help demonstrate this point. As a first example, differences in laboratory reporting may result in measurement error or misclassification. While the order for a particular laboratory assay is likely consistent within the healthcare system, patients frequently have a choice where to have that order fulfilled. Given the breadth of assays and reporting differences that may differ lab to lab [ 49 ], it is possible that the researcher working with the raw data may not consider all possible permutations. In other words, there may be lack of consistency in the reporting of the assay results. As a second example, raw clinical data requires interpretation to become actionable. A researcher interested in capturing a patient’s Charlson comorbidity index, which is based on 16 potential diagnoses plus the patient’s age [ 50 ], may never find such a variable in the EHR. Rather, this would require operationalization based on the raw data, each of which may be misclassified. Use of such composite measures introduces the notion of “differential item functioning”, whereby a summary indicator of a complexly measured health phenomenon may differ from group to group [ 51 ]. In this case, as opposed to a measurement error bias, this is one of residual confounding in that a key (unmeasured) variable is driving the differences. Remediation of these threats to validity may involve validation studies to determine the accuracy of a particular classifier, sensitivity analysis employing alternative interpretations when the raw data are available, and omitting or imputing biased or latent variables [ 40 , 41 , 52 ]. Importantly, in all cases, the epidemiologists should work with the various health care providers and personnel who have measured and recorded the data present in the EHR, as they likely understand it best.
Furthermore and related to “Billing versus Clinical versus Epidemiological Needs” section, the healthcare system in the U.S. is fragmented with multiple payers, both public and private, potentially exacerbating the data quality issues we describe, especially when linking data across healthcare systems. Single payer systems have enabled large and near-complete population-based studies due to data availability and consistency [ 53 , 54 , 55 ]. Data may also be inconsistent for retrospective longitudinal studies spanning many years if there have been changes to coding standards or practices over time, for example due to the transition from ICD-9 to ICD-10 largely occurring in the mid 2010s or the adoption of the Patient Protection and Affordable Care Act in the U.S. in 2010 with its accompanying changes in billing. Exploratory data analysis may reveal unexpected differences in key variables, by place or time, and recoding, when possible, can enforce consistency.
Sub-challenge #2.3: Unstructured data: clinical notes and reports
There may also be scenarios where structured data fields, while available, are not traditionally or consistently used within a given medical center or by a given provider. For example, reporting of adverse events of medications, disease symptoms, and vaccinations or hospitalizations occurring at different facility/health networks may not always be entered by providers in structured EHR fields. Instead, these types of patient experiences may be more likely to be documented in an unstructured clinical note, report (e.g. pathology or radiology report), or scanned document. Therefore, reliance on structured data to identify and study such issues may result in underestimation and potentially biased results.
Advances in NLP currently allow for information to be extracted from unstructured clinical notes and text fields in a reliable and accurate manner using computational methods. NLP utilizes a range of different statistical, machine learning, and linguistic techniques, and when applied to EHR data, has the potential to facilitate more accurate detection of events not traditionally located or consistently used in structured fields. Various NLP methods can be implemented in medical text analysis, ranging from simplistic and fast term recognition systems to more advanced, commercial NLP systems [ 56 ]. Several studies have successfully utilized text mining to extract information on a variety of health-related issues within clinical notes, such as opioid use [ 57 ], adverse events [ 58 , 59 ], symptoms (e.g., shortness of breath, depression, pain) [ 60 ], and disease phenotype information documented in pathology or radiology reports, including cancer stage, histology, and tumor grade [ 61 ], and lupus nephritis [ 32 ]. It is worth noting that scanned documents involve an additional layer of computation, relying on techniques such as optical character recognition, before NLP can be applied.
Hybrid approaches that combine both narrative and structured data, such as ICD codes, to improve accuracy of detecting phenotypes have also demonstrated high performance. Banerji et al. found that using ICD-9 codes to identify allergic drug reactions in the EHR had a positive predictive value of 46%, while an NLP algorithm in conjunction with ICD-9 codes resulted in a positive predictive value of 86%; negative predictive value also increased in the combined algorithm (76%) compared to ICD-9 codes alone (39%) [ 62 ]. In another example, researchers found that the combination of unstructured clinical notes with structured data for prediction tasks involving in-hospital mortality and 30-day hospital readmission outperformed models using either clinical notes or structured data alone [ 63 ]. As we move forward in analyzing EHR data, it will be important to take advantage of the wealth of information buried in unstructured data to assist in phenotyping patient characteristics and outcomes, capture missing confounders used in multivariate analyses, and develop prediction models.
Challenge #3: Missing measurements
While clinical notes may be useful to recover incomplete information from structured data fields, it may be the case that certain variables are not collected within the EHR at all. As mentioned above, it is important to remember that EHRs were not developed as a research tool (see “ Billing versus clinical versus epidemiological needs ” section), and important variables often used in epidemiologic research may not be typically included in EHRs including socioeconomic status (education, income, occupation) and SDOH [ 17 , 18 ]. Depending upon the interest of the provider or clinical importance placed upon a given variable, this information may be included in clinical notes. While NLP could be used to capture these variables, because they may not be consistently captured, there may be bias in identifying those with a positive mention as a positive case and those with no mention as a negative case. For example, if a given provider inquires about homelessness of a patient based on knowledge of the patient’s situation or other external factors and documents this in the clinical note, we have greater assurance that this is a true positive case. However, lack of mention of homelessness in a clinical note should not be assumed as a true negative case for several reasons: not all providers may feel comfortable asking about and/or documenting homelessness, they may not deem this variable worth noting, or implicit bias among clinicians may affect what is captured. As a result, such cases (i.e. no mention of homelessness) may be incorrectly identified as “not homeless,” leading to selection bias should a researcher form a cohort exclusively of patients who are identified as homeless in the EHR.
Not adjusting for certain measurements missing from EHR data can also lead to biased results if the measurement is an important confounder. Consider the example of distinguishing between prevalent and incident cases of disease when examining associations between disease treatments and patient outcomes [ 64 ]. The first date of an ICD code entered for a given patient may not necessarily be the true date of diagnosis, but rather documentation of an existing diagnosis. This limits the ability to adjust for disease duration, which may be an important confounder in studies comparing various treatments with patient outcomes over time, and may also lead to reverse causality if disease sequalae are assumed to be risk factors.
Methods to supplement EHR data with external data have been used to capture missing information. These methods may include imputation if information (e.g. race, lab values) is collected on a subset of patients within the EHR. It is important to examine whether missingness occurs completely at random or at random (“ignorable”), or not at random (“non-ignorable”), using the data available to determine factors associated with missingness, which will also inform the best imputation strategy to pursue, if any [ 65 , 66 ]. As an example, suppose we are interested in ascertaining a patient's BMI from the EHR. If men were less likely to have BMI measured than women, the probability of missing data (BMI) depends on the observed data (gender) and may therefore be predictable and imputable. On the other hand, suppose underweight individuals were less likely to have BMI measured; the probability of missing data depends on its own value, and as such is non-predictable and may require a validation study to confirm. Alternatively to imputing missing data, surrogate measures may be used, such as inferring area-based SES indicators, including median household income, percent poverty, or area deprivation index, by zip code [ 67 , 68 ]. Lastly, validation studies utilizing external datasets may prove helpful, such as supplementing EHR data with claims data that may be available for a subset of patients (see Challenge #4 ).
As EHRs are increasingly being used for research, there are active pushes to include more structured data fields that are important to population health research, such as SDOH [ 69 ]. Inclusion of such factors are likely to result in improved patient care and outcomes, through increased precision in disease diagnosis, more effective shared decision making, identification of risk factors, and tailoring services to a given population’s needs [ 70 ]. In fact, a recent review found that when individual level SDOH were included in predictive modeling, they overwhelmingly improved performance in medication adherence, risk of hospitalization, 30-day rehospitalizations, suicide attempts, and other healthcare services [ 71 ]. Whether or not these fields will be utilized after their inclusion in the EHR may ultimately depend upon federal and state incentives, as well as support from local stakeholders, and this does not address historic, retrospective analyses of these data.
Challenge #4: Missing visits
Beyond missing variable data that may not be captured during a clinical encounter, either through structured data or clinical notes, there also may be missing information for a patient as a whole. This can occur in a variety of ways; for example, a patient may have one or two documented visits in the EHR and then is never seen again (i.e. right censoring due to lost to follow-up), or a patient is referred from elsewhere to seek specialty care, with no information captured regarding other external issues (i.e. left censoring). This may be especially common in circumstances where a given EHR is more likely to capture specialty clinics versus primary care (see Challenge #1 ). A third scenario may include patients who appear, then are not observed for a long period of time, and then reappear: this case is particularly problematic as it may appear the patient was never lost to follow up but simply had fewer visits. In any of these scenarios, a researcher will lack a holistic view of the patient’s experiences, diagnoses, results, and more. As discussed above, assuming absence of a diagnostic code as absence of disease may lead to information and/or selection bias. Further, it has been demonstrated that one key source of bias in EHRs is “informed presence” bias, where those with more medical encounters are more likely to be diagnosed with various conditions (similar to Berkson’s bias) [ 72 ].
Several solutions to these issues have been proposed. For example, it is common for EHR studies to condition on observation time (i.e. ≥n visits required to be eligible into cohort); however, this may exclude a substantial amount of patients with certain characteristics, incurring a selection bias or limiting the generalizability of study findings (see Challenge #1 ). Other strategies attempt to account for missing visit biases through longitudinal imputation approaches; for example, if a patient missed a visit, a disease activity score can be imputed for that point in time, given other data points [ 73 , 74 ]. Surrogate measures may also be used to infer patient outcomes, such as controlling for “informative” missingness as an indicator variable or using actual number of missed visits that were scheduled as a proxy for external circumstances influencing care [ 20 ]. To address “informed presence” bias described above, conditioning on the number of health-care encounters may be appropriate [ 72 ]. Understanding the reason for the missing visit may help identify the best course of action and before imputing, one should be able to identify the type of missingness, whether “informative” or not [ 65 , 66 ]. For example, if distance to a healthcare location is related to appointment attendance, being able to account for this in analysis would be important: researchers have shown how the catchment of a healthcare facility can induce selection bias [ 21 ]. Relatedly, as telehealth becomes more common fueled by the COVID-19 pandemic [ 75 , 76 ], virtual visits may generate missingness of data recorded in the presence of a provider (e.g., blood pressure if the patient does not have access to a sphygmomanometer; see Challenge #3 ), or necessitate a stratified analysis by visit type to assess for effect modification.
Another common approach is to supplement EHR information with external data sources, such as insurance claims data, when available. Unlike a given EHR, claims data are able to capture a patient’s interaction with the health care system across organizations, and additionally includes pharmacy data such as if a prescription was filled or refilled. Often researchers examine a subset of patients eligible for Medicaid/Medicare and compare what is documented in claims with information available in the EHR [ 77 ]. That is, are there additional medications, diagnoses, hospitalizations found in the claims dataset that were not present in the EHR. In a study by Franklin et al., researchers utilized a linked database of Medicare Advantage claims and comprehensive EHR data from a multi-specialty outpatient practice to determine which dataset would be more accurate in predicting medication adherence [ 77 ]. They found that both datasets were comparable in identifying those with poor adherence, though each dataset incorporated different variables.
While validation studies such as those using claims data allow researchers to gain an understanding as to how accurate and complete a given EHR is, this may only be limited to the specific subpopulation examined (i.e. those eligible for Medicaid, or those over 65 years for Medicare). One study examined congruence between EHR of a community health center and Medicaid claims with respect to diabetes [ 78 ]. They found that patients who were older, male, Spanish-speaking, above the federal poverty level, or who had discontinuous insurance were more likely to have services documented in the EHR as compared to Medicaid claims data. Therefore, while claims data may help supplement and validate information in the EHR, on their own they may underestimate care in certain populations.
Research utilizing EHR data has undoubtedly positively impacted the field of public health through its ability to provide large-scale, longitudinal data on a diverse set of patients, and will continue to do so in the future as more epidemiologists take advantage of this data source. EHR data’s ability to capture individuals that traditionally aren’t included in clinical trials, cohort studies, and even claims datasets allows researchers to measure longitudinal outcomes in patients and perhaps change the understanding of potential risk factors.
However, as outlined in this review, there are important caveats to EHR analysis that need to be taken into account; failure to do so may threaten study validity. The representativeness of EHR data depends on the catchment area of the center and corresponding target population. Tools are available to evaluate and remedy these issues, which are critical to study validity as well as extrapolation of study findings. Data availability and interpretation, missing measurements, and missing visits are also key challenges, as EHRs were not specifically developed for research purposes, despite their common use for such. Taking advantage of all available EHR data, whether it be structured or unstructured fields through NLP, will be important in understanding the patient experience and identifying key phenotypes. Beyond methods to address these concerns, it will remain crucial for epidemiologists and data analysts to engage with clinicians and informaticians at their institutions to ensure data quality and accessibility by forming multidisciplinary teams around specific research projects. Lastly, integration across multiple EHRs, or datasets that encompass multi-institutional EHR records, add an additional layer of data quality and validity issues, with the potential to exacerbate the above-stated challenges found within a single EHR. At minimum, such studies should account for correlated errors [ 79 , 80 ], and investigate whether modularization, or submechanisms that determine whether data are observed or missing in each EHR, exist [ 65 ].
The identified challenges may also apply to secondary analysis of other large healthcare databases, such as claims data, although it is important not to conflate the two types of data. EHR data are driven by clinical care and claims data are driven by the reimbursement process where there is a financial incentive to capture diagnoses, procedures, and medications [ 48 ]. The source of data likely influences the availability, accuracy, and completeness of data. The fundamental representation of data may also differ as a record in a claims database corresponds to a “claim” as opposed to an “encounter” in the EHR. As such, the representativeness of the database populations, the sensitivity and specificity of variables, as well as the mechanisms of missingness in claims data may differ from EHR data. One study that evaluated pediatric quality care measures, such as BMI, noted inferior sensitivity based on claims data alone [ 81 ]. Linking claims data to EHR data has been proposed to enhance study validity, but many of the caveats raised in herein still apply [ 82 ].
Although we focused on epidemiological challenges related to study validity, there are other important considerations for researchers working with EHR data. Privacy and security of data as well as institutional review board (IRB) or ethics board oversight of EHR-based studies should not be taken for granted. For researchers in the U.S., Goldstein and Sarwate described Health Insurance Portability and Accountability Act (HIPAA)-compliant approaches to ensure the privacy and security of EHR data used in epidemiological research, and presented emerging approaches to analyses that separate the data from analysis [ 83 ]. The IRB oversees the data collection process for EHR-based research and through the HIPAA Privacy Rule these data typically do not require informed consent provided they are retrospective and reside at the EHR’s institution [ 84 ]. Such research will also likely receive an exempt IRB review provided subjects are non-identifiable.
Conclusions
As EHRs are increasingly being used for research, epidemiologists can take advantage of the many tools and methods that already exist and apply them to the key challenges described above. By being aware of the limitations that the data present and proactively addressing them, EHR studies will be more robust, informative, and important to the understanding of health and disease in the population.
Availability of data and materials
All data and materials used in this review are described herein.
Abbreviations
Body Mass Index
Electronic Health Record
International Classification of Diseases
Institutional review board/ethics board
Health Insurance Portability and Accountability Act
Natural Language Processing
Social Determinants of Health
Socioeconomic Status
Adler-Milstein J, Holmgren AJ, Kralovec P, et al. Electronic health record adoption in US hospitals: the emergence of a digital “advanced use” divide. J Am Med Inform Assoc. 2017;24(6):1142–8.
Article PubMed PubMed Central Google Scholar
Office of the National Coordinator for Health Information Technology. ‘Office-based physician electronic health record adoption’, Health IT quick-stat #50. dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php . Accessed 15 Jan 2019.
Cowie MR, Blomster JI, Curtis LH, et al. Electronic health records to facilitate clinical research. Clin Res Cardiol. 2017;106(1):1–9.
Article PubMed Google Scholar
Casey JA, Schwartz BS, Stewart WF, et al. Using electronic health records for population health research: a review of methods and applications. Annu Rev Public Health. 2016;37:61–81.
Verheij RA, Curcin V, Delaney BC, et al. Possible sources of bias in primary care electronic health record data use and reuse. J Med Internet Res. 2018;20(5):e185.
Ni K, Chu H, Zeng L, et al. Barriers and facilitators to data quality of electronic health records used for clinical research in China: a qualitative study. BMJ Open. 2019;9(7):e029314.
Coleman N, Halas G, Peeler W, et al. From patient care to research: a validation study examining the factors contributing to data quality in a primary care electronic medical record database. BMC Fam Pract. 2015;16:11.
Kruse CS, Stein A, Thomas H, et al. The use of electronic health records to support population health: a systematic review of the literature. J Med Syst. 2018;42(11):214.
Shortreed SM, Cook AJ, Coley RY, et al. Challenges and opportunities for using big health care data to advance medical science and public health. Am J Epidemiol. 2019;188(5):851–61.
In: Smedley BD, Stith AY, Nelson AR, editors. Unequal treatment: confronting racial and ethnic disparities in health care. Washington (DC) 2003.
Chaudhry B, Wang J, Wu S, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med. 2006;144(10):742–52.
Cutler DM, Scott Morton F. Hospitals, market share, and consolidation. JAMA. 2013;310(18):1964–70.
Article CAS PubMed Google Scholar
Cocoros NM, Kirby C, Zambarano B, et al. RiskScape: a data visualization and aggregation platform for public health surveillance using routine electronic health record data. Am J Public Health. 2021;111(2):269–76.
Vader DT, Weldie C, Welles SL, et al. Hospital-acquired Clostridioides difficile infection among patients at an urban safety-net hospital in Philadelphia: demographics, neighborhood deprivation, and the transferability of national statistics. Infect Control Hosp Epidemiol. 2020;42:1–7.
Google Scholar
Dixon BE, Gibson PJ, Frederickson Comer K, et al. Measuring population health using electronic health records: exploring biases and representativeness in a community health information exchange. Stud Health Technol Inform. 2015;216:1009.
PubMed Google Scholar
Hernán MA, VanderWeele TJ. Compound treatments and transportability of causal inference. Epidemiology. 2011;22(3):368–77.
Casey JA, Pollak J, Glymour MM, et al. Measures of SES for electronic health record-based research. Am J Prev Med. 2018;54(3):430–9.
Polubriaginof FCG, Ryan P, Salmasian H, et al. Challenges with quality of race and ethnicity data in observational databases. J Am Med Inform Assoc. 2019;26(8-9):730–6.
U.S. Census Bureau. Health. Available at: https://www.census.gov/topics/health.html . Accessed 19 Jan 2021.
Gianfrancesco MA, McCulloch CE, Trupin L, et al. Reweighting to address nonparticipation and missing data bias in a longitudinal electronic health record study. Ann Epidemiol. 2020;50:48–51 e2.
Goldstein ND, Kahal D, Testa K, Burstyn I. Inverse probability weighting for selection bias in a Delaware community health center electronic medical record study of community deprivation and hepatitis C prevalence. Ann Epidemiol. 2021;60:1–7.
Gelman A, Lax J, Phillips J, et al. Using multilevel regression and poststratification to estimate dynamic public opinion. Unpublished manuscript, Columbia University. 2016 Sep 11. Available at: http://www.stat.columbia.edu/~gelman/research/unpublished/MRT(1).pdf . Accessed 22 Jan 2021.
Quick H, Terloyeva D, Wu Y, et al. Trends in tract-level prevalence of obesity in philadelphia by race-ethnicity, space, and time. Epidemiology. 2020;31(1):15–21.
Lesko CR, Buchanan AL, Westreich D, Edwards JK, Hudgens MG, Cole SR. Generalizing study results: a potential outcomes perspective. Epidemiology. 2017;28(4):553–61.
Westreich D, Edwards JK, Lesko CR, Stuart E, Cole SR. Transportability of trial results using inverse odds of sampling weights. Am J Epidemiol. 2017;186(8):1010–4.
Congressional Research Services (CRS). The Health Information Technology for Economic and Clinical Health (HITECH) Act. 2009. Available at: https://crsreports.congress.gov/product/pdf/R/R40161/9 . Accessed Jan 22 2021.
Hersh WR. The electronic medical record: Promises and problems. Journal of the American Society for Information Science. 1995;46(10):772–6.
Article Google Scholar
Collecting sexual orientation and gender identity data in electronic health records: workshop summary. Washington (DC) 2013.
Committee on the Recommended Social and Behavioral Domains and Measures for Electronic Health Records; Board on Population Health and Public Health Practice; Institute of Medicine. Capturing social and behavioral domains and measures in electronic health records: phase 2. Washington (DC): National Academies Press (US); 2015.
Goff SL, Pekow PS, Markenson G, et al. Validity of using ICD-9-CM codes to identify selected categories of obstetric complications, procedures and co-morbidities. Paediatr Perinat Epidemiol. 2012;26(5):421–9.
Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. J Clin Epidemiol. 2005;58(4):323–37.
Gianfrancesco MA. Application of text mining methods to identify lupus nephritis from electronic health records. Lupus Science & Medicine. 2019;6:A142.
National Library of Medicine. SNOMED CT to ICD-10-CM Map. Available at: https://www.nlm.nih.gov/research/umls/mapping_projects/snomedct_to_icd10cm.html . Accessed 2 Jul 2021.
Klabunde CN, Harlan LC, Warren JL. Data sources for measuring comorbidity: a comparison of hospital records and medicare claims for cancer patients. Med Care. 2006;44(10):921–8.
Burles K, Innes G, Senior K, Lang E, McRae A. Limitations of pulmonary embolism ICD-10 codes in emergency department administrative data: let the buyer beware. BMC Med Res Methodol. 2017;17(1):89.
Asgari MM, Wu JJ, Gelfand JM, Salman C, Curtis JR, Harrold LR, et al. Validity of diagnostic codes and prevalence of psoriasis and psoriatic arthritis in a managed care population, 1996-2009. Pharmacoepidemiol Drug Saf. 2013;22(8):842–9.
Hoffman S, Podgurski A. Big bad data: law, public health, and biomedical databases. J Law Med Ethics. 2013;41(Suppl 1):56–60.
Adler-Milstein J, Jha AK. Electronic health records: the authors reply. Health Aff. 2014;33(10):1877.
Geruso M, Layton T. Upcoding: evidence from medicare on squishy risk adjustment. J Polit Econ. 2020;12(3):984–1026.
Lash TL, Fox MP, Fink AK. Applying quantitative bias analysis to epidemiologic data. New York: Springer-Verlag New York; 2009.
Book Google Scholar
Gustafson P. Measurement error and misclassification in statistics and epidemiology: impacts and Bayesian adjustments. Boca Raton: Chapman and Hall/CRC; 2004.
Duda SN, Shepherd BE, Gadd CS, et al. Measuring the quality of observational study data in an international HIV research network. PLoS One. 2012;7(4):e33908.
Article CAS PubMed PubMed Central Google Scholar
Shepherd BE, Yu C. Accounting for data errors discovered from an audit in multiple linear regression. Biometrics. 2011;67(3):1083–91.
Weiskopf NG, Hripcsak G, Swaminathan S, et al. Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform. 2013;46(5):830–6.
Kaiser Health News. As coronavirus strikes, crucial data in electronic health records hard to harvest. Available at: https://khn.org/news/as-coronavirus-strikes-crucial-data-in-electronic-health-records-hard-to-harvest/ . Accessed 15 Jan 2021.
Reeves JJ, Hollandsworth HM, Torriani FJ, Taplitz R, Abeles S, Tai-Seale M, et al. Rapid response to COVID-19: health informatics support for outbreak management in an academic health system. J Am Med Inform Assoc. 2020;27(6):853–9.
Grange ES, Neil EJ, Stoffel M, Singh AP, Tseng E, Resco-Summers K, et al. Responding to COVID-19: The UW medicine information technology services experience. Appl Clin Inform. 2020;11(2):265–75.
Madigan D, Ryan PB, Schuemie M, et al. Evaluating the impact of database heterogeneity on observational study results. Am J Epidemiol. 2013;178(4):645–51.
Lippi G, Mattiuzzi C. Critical laboratory values communication: summary recommendations from available guidelines. Ann Transl Med. 2016;4(20):400.
Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–83.
Jones RN. Differential item functioning and its relevance to epidemiology. Curr Epidemiol Rep. 2019;6:174–83.
Edwards JK, Cole SR, Troester MA, Richardson DB. Accounting for misclassified outcomes in binary regression models using multiple imputation with internal validation data. Am J Epidemiol. 2013;177(9):904–12.
Satkunasivam R, Klaassen Z, Ravi B, Fok KH, Menser T, Kash B, et al. Relation between surgeon age and postoperative outcomes: a population-based cohort study. CMAJ. 2020;192(15):E385–92.
Melamed N, Asztalos E, Murphy K, Zaltz A, Redelmeier D, Shah BR, et al. Neurodevelopmental disorders among term infants exposed to antenatal corticosteroids during pregnancy: a population-based study. BMJ Open. 2019;9(9):e031197.
Kao LT, Lee HC, Lin HC, Tsai MC, Chung SD. Healthcare service utilization by patients with obstructive sleep apnea: a population-based study. PLoS One. 2015;10(9):e0137459.
Article PubMed PubMed Central CAS Google Scholar
Jung K, LePendu P, Iyer S, Bauer-Mehren A, Percha B, Shah NH. Functional evaluation of out-of-the-box text-mining tools for data-mining tasks. J Am Med Inform Assoc. 2015;22(1):121–31.
Canan C, Polinski JM, Alexander GC, et al. Automatable algorithms to identify nonmedical opioid use using electronic data: a systematic review. J Am Med Inform Assoc. 2017;24(6):1204–10.
Iqbal E, Mallah R, Jackson RG, et al. Identification of adverse drug events from free text electronic patient records and information in a large mental health case register. PLoS One. 2015;10(8):e0134208.
Rochefort CM, Verma AD, Eguale T, et al. A novel method of adverse event detection can accurately identify venous thromboembolisms (VTEs) from narrative electronic health record data. J Am Med Inform Assoc. 2015;22(1):155–65.
Koleck TA, Dreisbach C, Bourne PE, et al. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc. 2019;26(4):364–79.
Wang L, Luo L, Wang Y, et al. Natural language processing for populating lung cancer clinical research data. BMC Med Inform Decis Mak. 2019;19(Suppl 5):239.
Banerji A, Lai KH, Li Y, et al. Natural language processing combined with ICD-9-CM codes as a novel method to study the epidemiology of allergic drug reactions. J Allergy Clin Immunol Pract. 2020;8(3):1032–1038.e1.
Zhang D, Yin C, Zeng J, et al. Combining structured and unstructured data for predictive models: a deep learning approach. BMC Med Inform Decis Mak. 2020;20(1):280.
Farmer R, Mathur R, Bhaskaran K, Eastwood SV, Chaturvedi N, Smeeth L. Promises and pitfalls of electronic health record analysis. Diabetologia. 2018;61:1241–8.
Haneuse S, Arterburn D, Daniels MJ. Assessing missing data assumptions in EHR-based studies: a complex and underappreciated task. JAMA Netw Open. 2021;4(2):e210184.
Groenwold RHH. Informative missingness in electronic health record systems: the curse of knowing. Diagn Progn Res. 2020;4:8.
Berkowitz SA, Traore CY, Singer DE, et al. Evaluating area-based socioeconomic status indicators for monitoring disparities within health care systems: results from a primary care network. Health Serv Res. 2015;50(2):398–417.
Kind AJH, Buckingham WR. Making neighborhood-disadvantage metrics accessible - the neighborhood atlas. N Engl J Med. 2018;378(26):2456–8.
Cantor MN, Thorpe L. Integrating data on social determinants of health into electronic health records. Health Aff. 2018;37(4):585–90.
Adler NE, Stead WW. Patients in context--EHR capture of social and behavioral determinants of health. N Engl J Med. 2015;372(8):698–701.
Chen M, Tan X, Padman R. Social determinants of health in electronic health records and their impact on analysis and risk prediction: a systematic review. J Am Med Inform Assoc. 2020;27(11):1764–73.
Goldstein BA, Bhavsar NA, Phelan M, et al. Controlling for informed presence bias due to the number of health encounters in an electronic health record. Am J Epidemiol. 2016;184(11):847–55.
Petersen I, Welch CA, Nazareth I, et al. Health indicator recording in UK primary care electronic health records: key implications for handling missing data. Clin Epidemiol. 2019;11:157–67.
Li R, Chen Y, Moore JH. Integration of genetic and clinical information to improve imputation of data missing from electronic health records. J Am Med Inform Assoc. 2019;26(10):1056–63.
Koonin LM, Hoots B, Tsang CA, Leroy Z, Farris K, Jolly T, et al. Trends in the use of telehealth during the emergence of the COVID-19 pandemic - United States, January-March 2020. MMWR Morb Mortal Wkly Rep. 2020;69(43):1595–9.
Barnett ML, Ray KN, Souza J, Mehrotra A. Trends in telemedicine use in a large commercially insured population, 2005-2017. JAMA. 2018;320(20):2147–9.
Franklin JM, Gopalakrishnan C, Krumme AA, et al. The relative benefits of claims and electronic health record data for predicting medication adherence trajectory. Am Heart J. 2018;197:153–62.
Devoe JE, Gold R, McIntire P, et al. Electronic health records vs Medicaid claims: completeness of diabetes preventive care data in community health centers. Ann Fam Med. 2011;9(4):351–8.
Schmajuk G, Li J, Evans M, Anastasiou C, Izadi Z, Kay JL, et al. RISE registry reveals potential gaps in medication safety for new users of biologics and targeted synthetic DMARDs. Semin Arthritis Rheum. 2020 Dec;50(6):1542–8.
Izadi Z, Schmajuk G, Gianfrancesco M, Subash M, Evans M, Trupin L, et al. Rheumatology Informatics System for Effectiveness (RISE) practices see significant gains in rheumatoid arthritis quality measures. Arthritis Care Res. 2020. https://doi.org/10.1002/acr.24444 .
Angier H, Gold R, Gallia C, Casciato A, Tillotson CJ, Marino M, et al. Variation in outcomes of quality measurement by data source. Pediatrics. 2014;133(6):e1676–82.
Lin KJ, Schneeweiss S. Considerations for the analysis of longitudinal electronic health records linked to claims data to study the effectiveness and safety of drugs. Clin Pharmacol Ther. 2016;100(2):147–59.
Goldstein ND, Sarwate AD. Privacy, security, and the public health researcher in the era of electronic health record research. Online J Public Health Inform. 2016;8(3):e207.
U.S. Department of Health and Human Services (HHS). 45 CFR 46. http://www.hhs.gov/ohrp/regulations-and-policy/regulations/45-cfr-46/index.html .
Download references
Acknowledgements
The authors thank Dr. Annemarie Hirsch, Department of Population Health Sciences, Geisinger, for assistance in conceptualizing an earlier version of this work.
Research reported in this publication was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number K01AR075085 (to MAG) and the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health under Award Number K01AI143356 (to NDG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and affiliations.
Division of Rheumatology, University of California School of Medicine, San Francisco, CA, USA
Milena A. Gianfrancesco
Department of Epidemiology and Biostatistics, Drexel University Dornsife School of Public Health, 3215 Market St., Philadelphia, PA, 19104, USA
Neal D. Goldstein
You can also search for this author in PubMed Google Scholar
Contributions
Both authors conceptualized, wrote, and approved the final submitted version.
Corresponding author
Correspondence to Neal D. Goldstein .
Ethics declarations
Ethics approval and consent to participate.
Not applicable
Consent for publication
Competing interests.
The authors have no competing interests to declare
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.

Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Gianfrancesco, M.A., Goldstein, N.D. A narrative review on the validity of electronic health record-based research in epidemiology. BMC Med Res Methodol 21 , 234 (2021). https://doi.org/10.1186/s12874-021-01416-5
Download citation
Received : 02 July 2021
Accepted : 28 September 2021
Published : 27 October 2021
DOI : https://doi.org/10.1186/s12874-021-01416-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Electronic health records
- Data quality
- Secondary analysis
BMC Medical Research Methodology
ISSN: 1471-2288
- General enquiries: [email protected]
- Research article
- Open access
- Published: 02 March 2020
A systematic review of patient access to medical records in the acute setting: practicalities, perspectives and ethical consequences
- Stephanie N. D’Costa 1 ,
- Isla L. Kuhn 2 &
- Zoë Fritz ORCID: orcid.org/0000-0001-9403-409X 2
BMC Medical Ethics volume 21 , Article number: 18 ( 2020 ) Cite this article
13k Accesses
24 Citations
11 Altmetric
Metrics details
Internationally, patient access to notes is increasing. This has been driven by respect for patient autonomy, often recognised as a primary tenet of medical ethics: patients should be able to access their records to be fully engaged with their care. While research has been conducted on the impact of patient access to outpatient and primary care records and to patient portals, there is no such review looking at access to hospital medical records in real time, nor an ethical analysis of the issues involved in such a change in process.
This study employed a systematic review framework in two stems, to integrate literature identified from two searches: Medline, CINAHL and Scopus databases were conducted, (for (1) hospitalised patients, patient access to records and its effects on communication and trust within the doctor-patient relationship; and (2) patient access to medical records and the ethical implications identified). The qualitative and quantitative results of both searches were integrated and critically analysed.
3954 empirical and 4929 ethical studies were identified; 18 papers representing 16 studies were identified for review (12 empirical and 6 ethical). The review reveals a consensus that our current approach to giving information to patients – almost exclusively verbally – is insufficient; that patient access to notes is a welcome next step for patient-centred care, but that simply allowing full access, without explanation or summary, is also insufficient. Several ethical implications need to be considered: increased information could improve patient trust and knowledge but might transfer an (unwelcome) sense of responsibility to patients; doctors and patients have conflicting views on how much information should be shared and when; sharing written information might increase the already significant disparity in access to health care, and have unforeseen opportunity costs. The impact on medical practice of sharing notes in real time will also need to be evaluated.
Conclusions
The review presents encouraging data to support patient access to medical notes. However, sharing information is a critical part of clinical practice; changing how it is done could have significant empirical and ethical impacts; any changes should be carefully evaluated.
Peer Review reports
It is unusual for patients to request access to their medical hospital records, despite their legal right to do so [ 1 ]. The U.K. government mandated that patients should be able to readily access their electronic medical record by 2018, a promise which has not been fulfilled, mostly due to logistical difficulties [ 2 ]. This mandate was built on respect for patient autonomy as a primary tenet of medical ethics: patients should be able to access their records to be fully engaged with their care. Access to records allows patients to be more informed which may increase opportunities for them to question their care plans and request second opinions.
Internationally, patients are more readily able to access their notes, and there has been evidence of positive outcomes in maternity records [ 3 ]; in primary care [ 4 , 5 ]; for specific diseases, [ 6 , 7 ] and for specific interventions [ 8 , 9 ]. A 2003 (Ross and Lin) [ 10 ] and 2007 (Ferreira et al) [ 11 ] review of the literature in these fields found that patient access was unlikely to cause harm and can improve doctor-patient communication and relations; the latter review also identified the potential for patients to spot and correct mistakes in their records.
More recently, the use of patient ‘portals’ – an electronic route to targeted parts of the medical record – has become more common. Several systematic reviews on the design, use and impact of such portals have been conducted [ 12 , 13 , 14 ]. Patients are generally enthusiastic about the possibility of accessibility, and positive or neutral health outcomes were observed. However, it was noted that clinician contact for portal users increased, and, perhaps related to this, disparity of uptake among different ethnic and socioeconomic groups was noted.
While these reviews demonstrate significant bodies of research on the impact of patient access to outpatient and primary care records and to patient portals (see Table 1 for a summary table of the systematic reviews in these domains), there is no such review looking at access to hospital medical records in real time, nor an ethical analysis of the issues involved in such a change in process.
Real-time access to medical records (particularly as they are currently written) may have unintended consequences on patient care both directly and indirectly – for example, by altering how things are recorded in the notes.
In this paper, we focus on adult access to notes in the medical acute care setting. We define this as the environment which comprises an adult medical patient’s presentation to hospital and their initial (up to 5 days) in-patient stay. This is a busy environment in which a sick patient generally only receives verbal communication, and in which decisions need to be made quickly, often by or with clinicians unfamiliar to the patient. In this context, access to notes may serve a different purpose than in the chronic disease or outpatient setting. Our aim was to review empirical papers relating to patient access and contribution to medical records, and consider the ethical issues raised by this proposed change in practice to fully appreciate the consequences of access to notes in real time.
Our review therefore set out to answer two questions:
1) What studies have there been of sharing records with medical patients in the in-patient setting, and in particular on the impact on trust and communication between patients and doctors?
2) What are the ethical issues associated with sharing records with medical patients?
This study employed a systematic review framework in two stems, to integrate literature identified from two searches surrounding our research questions. This ensured a robust, replicable searching strategy from which we could extract data clearly defined by inclusion and exclusion criteria (an initial attempt to search ethical issues relating to sharing medical records in acute care yielded no relevant results). We conducted critical interpretive synthesis [ 16 ] to the data extracted, an application of qualitative enquiry that allowed us to critically analyse and integrate both the qualitative and quantitative results of both searches into main themes.
The review was registered on the PROSPERO database (registration ID CRD42018114125). PRISMA guidelines have been used to inform the methodology and write up.
Identification of studies
A replicable search strategy was developed to answer our two research questions, using two literature searches, on the Medline via OVID, CINAHL via Ebsco and Scopus databases (See Appendix 1 for the full search strategies for both searches). Searches were run on the 23rd February 2018. Reference lists of included studies were reviewed for additional papers. A complete record of all identified articles was kept on a managed reference database.
Literature search of the empirical data
Search words, phrases and subject headings (including MeSH) were used to search for literature surrounding the topics of (1) hospitalised patients, (2) patient access to records and (3) its effects on communication and trust within the doctor-patient relationship.
The inclusion criteria limited the literature to studies about adult, hospitalised patients in the acute setting. Limits were applied for English language papers published since 1997 were included. Exclusion criteria consisted of paediatric, disease-specific studies and those focussed on confidentiality and data sharing. Studies relating to the design of a system allowing patient access to records were also excluded.
Literature search of the ethical issues
The second search consisted of a range of terms for (1) patient access to records and (2) ethical implications. This search therefore did not specify hospitalised patients or the effect access to notes has on communication and trust and was run from inception to the search date. The exclusion criteria remained the same.
Study selection
For each search, the titles and abstracts of references were screened by one reviewer (either SD or ZF) who selected those appropriate for full text analysis. 100 references in every 1000 were independently screened by both reviewers to assess for concordance and prevent drift, refining the inclusion criteria if needed. Any references where there was ambiguity were discussed by both authors and a decision made. Reference lists of included studies were screened by both authors. The results of the study selection are shown in Fig. 1 .
PRISMA Article Selection Flow chart
Data extraction and risk of bias
SD extracted the following data from the included studies: setting, nature of study, sample size, nature and contribution of participants, nature of analysis and summary of results, shown in Table 1 . Both researchers conducted thematic analysis on the papers, identifying four major themes.
Planned methods of analysis
Meta-analysis was inappropriate for the heterogenous nature of the search results and therefore a critical interpretive synthesis [ 16 ] was undertaken to discover emerging themes from the literature. Analysis of the papers was followed by extraction of data and discussion between the two authors, to consider the themes underlying these results. The ethics literature, which encompassed a wider range of settings than the empirical literature, was examined for themes which would be applicable across health care settings. An iterative process was utilised, examining and grouping them into overarching themes that both organised and illustrated the findings of the review.
Of the 3954 empirical and 4929 ethical studies identified through the two searches, 18 papers representing 16 studies were identified for review (12 empirical and 6 ethical) see Fig. 1 .
Two studies used questionnaires [ 17 , 18 ]; four used interviews or focus groups [ 19 , 20 , 21 , 22 , 23 , 24 ]; two used mixed methods [ 25 , 26 ]. One note analysis, [ 27 ] one portal analysis [ 28 ] and one clinical trial [ 29 , 30 ] was conducted, and six analysis articles were identified [ 31 , 32 , 33 , 34 , 35 , 36 ]. One empirical study came from each of Israel [ 19 ], Norway [ 20 , 21 ] and Canada [ 18 ]; the rest originated from the USA. No papers looked at perspectives of the multidisciplinary team. The data extraction is summarised in Table 2 .
Four main themes emerged on analysis: Impact on patient care; Conflicts between patient and physician perspective; divergent views on doctor and patient roles; cultural differences and societal risks.
Impact on patient care
Sharing notes was seen to empower patients by improving trust and knowledge [ 30 ], facilitating patients to work with doctors [ 28 ]. Communication of written information was considered superior to verbal explanations; one patient was reported as saying “Yeah, they come and update me but..I mean I can’t keep track of it all. That’s why I like this.” [ 24 ] No studies revealed objective changes in care such as reduced length of stay. Access to their own notes might enable patients to correct inaccuracies, [ 21 , 36 ] although this raised the possibility of patients feeling responsible if something was missed: [ 20 , 32 ]
“patients could end up feeling they are to blame for their own poor outcomes.” [ 32 ]
Some participants thought written information might ‘facilitate verbal communication’. [ 26 ] Others were concerned that a written note might supplant face-to-face interaction [ 22 ]; this did not manifest in the only study to trial giving patients a written daily summary [ 17 ].
Conflict between doctor and patient perspectives
Patients and doctors had discordant perceptions of how accessing the medical record might affect care: whilst doctors were concerned access to notes will overwhelm or unnecessarily worry patients, [ 17 , 24 ] patients were reassured by the shared information [ 37 ]. Grossman et al suggested that ‘it may be prudent to omit or explain potentially alarming information that carries a low degree of certainty such as a cancer on a differential diagnosis list” [ 28 ] .
A reoccurring conflict was the release of lab (and other) results in real time – patients strongly supported this whereas doctors preferred a delay, [ 24 , 29 , 31 ] in part so they could interpret them appropriately, offer support and create a future healthcare plan. Without this, some participants theorised that results could be prone to misinterpretation and unnecessary anxiety could be provoked [ 32 ]. As a physician participant said: “one of the primary duties of a physician is not only to alert the patient to abnormal results but also to educate them on their condition and appraise them of the follow up that will be needed” [ 32 ] . If delayed release did exist, however, there was a question about who would take responsibility for this [ 31 ].. Interestingly, this was not mentioned in the papers reporting direct experience.
There was also debate about whether patients should be co-creators of notes: Doctors, again hypothesising, were concerned that patients editing their own record might make them less reliable [ 34 , 35 ].
Divergent views on doctor and patient roles
A range of alternative approaches have been developed to share non-verbal information, and they reveal a variety of implicit perspectives about the role of the patient and the doctor. Tools designed to ensure patient choice and satisfaction are for those who perceive the patient as client ; one participant was quoted as saying: “I would like to be able to see background information [ about my doctor] like where they went to school” [ 25 ]. Providing information in the hope that patients will become more actively involved in their care see the patient as collaborator [ 22 , 29 ]. The different perspectives influence the purpose (and extent) of information sharing.
Cultural difference and societal risks
Different healthcare systems worldwide vary in their approach and concerns regarding access to notes – one study set in Israel found that the doctors more willing to share notes with patients originated from English-speaking countries, suggesting a cultural influence towards this [ 19 ]. In some countries such as the USA and Norway, liability seems to be more of a concern for the doctors and more of a motive for patients to access notes [ 21 , 24 ].
Across geographical boundaries, however, there was a recognition that there would be variation in patients’ willingness and ability to access notes, and that this might lead to disparity in health care, [ 22 , 35 ] with those from lower socioeconomic groups less likely to engage despite an often greater need; ‘{to] what extent should less engaged individuals be punished for their ‘ignorance’ [ 35 ] . As Lyles et al stated : “there is an ethical imperative to work to reduce the potential for the emergence or amplification of health disparities with respect to portal use’ [ 33 ] . Large screens, simple formats and buttons will help accessibility for some [ 26 , 38 ]; empirical research assessing the impact on access to health care or impact on different socioeconomic groups was not identified.
Finally, the questions of privacy and security of patient notes were raised, although papers focussing solely on this issue were excluded from the study. Some patients were concerned about the security of having information on their own devices, [ 26 ] while others did not voice privacy concerns [ 25 ]. Patients need to be able to trust their details are stored and shared securely, so they can contribute to them in a transparent manner [ 35 ].
The review reveals a consensus that our current approach to giving information to patients – almost exclusively verbally – is insufficient; that patient access to notes is a welcome next step for patient-centred care, but that simply allowing full access, without explanation or summary, is also insufficient. Several ethical implications need to be considered: increased information could improve patient trust and knowledge but might transfer an (unwelcome) sense of responsibility to patients; doctors and patients have conflicting views on how much information should be shared and when; sharing written information might increase the already significant disparity in access to health care, and have unforeseen opportunity costs.
It is also clear that we need to consider the impact that sharing notes in real time will have on medical practice.
Trust and the medical record
Although trust, both in doctors individually and generally, is often measured, it is rarely sufficiently specified in the medical literature. Trust is necessary when there is a degree of uncertainty and vulnerability (Becker 1996), both of which are present in the patient-doctor relationship; uncertainty about diagnosis and treatment, vulnerability not only because the patient is physically unwell, but because of the anxiety which often accompanies illness, and which can affect judgment. Trust is often described as a ‘three place relation’: ‘A’ trusts ‘B’ with ‘C’ [ 39 ].
In healthcare, the factors which can determine trust can relate to the patient (‘A’) and the doctor (‘B’), as well as what is entrusted (‘C’), namely the patient’s care [ 40 ]. Since the degree of care required is related to the severity and circumstances of the illness, these are also factors which can affect the patient’s vulnerability and need to trust. While trust is necessary for a functioning patient-doctor relationship, too much trust could be detrimental [ 41 ]. It may lead to reduced patient involvement in decision-making, or fewer questions being asked, leading to the possibility of sub-standard patient care.
What we want to achieve is well-placed patient trust, a concept O’Neil refers to as trust of the trustworthy, [ 42 ] where a patient can be confident that their trust in their clinician is justified and thus can reasonably entrust decisions and actions about their care to him or her. This places an obligation on clinicians to be trustworthy, but it also requires patients to be able to ask questions to satisfy themselves that their trust is well-placed. Providing access to medical records enables patients to determine what they are entrusting (more about what is wrong with them, and more about what treatments and investigations are planned) and enables them to place their trust well (or withhold it). Patients reading their own records might in turn alter physicians’ behaviours to be more trustworthy: they may, as they have done with clinic letters, modulate their language and ensure better verbal communication to avoid misconstruction of what is written.
Increased knowledge, increased responsibility?
While trust is important, the relationship between trust and autonomy has been well explored [ 43 ]. In medical ethics analysis of the last 40 years, autonomy has been given primacy [ 44 , 45 ]; part of respecting patient autonomy is ensuring that they have sufficient information to participate in shared decision-making [ 46 ]. There appears to be a recognition that the current approach – of only relaying verbal information to patients until their discharge – is inadequate. Patients forget, [ 47 ] relatives are concerned, questions are not asked [ 48 ].
It is thus unsurprising that imparting more (or more accessible) information to patients was welcomed by both patients and doctors. However, concerns were expressed that giving more information to patients also transferred responsibility to them: responsibility to check for errors; to deal with uncertainty; to worry about results. This responsibility may not always be desired by the patient. As Alfred Taubert says: “In the so-called co-operative mode, guidance dominates to the point where most patients, realistically and appropriately, want the doctor to take responsibility for their health.” By giving patients increased information, we may be removing their choice to defer responsibility – and associated ‘emotional work’ [ 49 ] or worry - to their physician.
Too much information, too soon?
A specific example of emotional work or worry related to receiving test results in real time: whilst patients expressed a strong desire for this, doctors’ concerns are two-fold. Firstly, they were concerned that patients lack the medical expertise to gauge the clinical importance of results. Secondly, they were worried that they (the doctor) would not be present to offer support and interpretation if the patient receives distressing news. Receiving emotional support from their doctor was a primary reason found for why patients audio-record consultations [ 50 ]; getting results without the doctor present would deprive them of that immediate support. Outside the acute setting, Milliat-Guittard showed that 21% of breast cancer patients did not want to hold records; they did not want to come across a comment that they were not expecting. Instead, they wished to come to terms with the disease in their own way [ 51 ].
Unintended worsening of inequality
Some interventions unintentionally increase inequalities by disproportionately benefiting less disadvantaged groups [ 52 ]. Giving patients access to records might be one such intervention: clinical teams acknowledged that they were working in a stretched system - an intervention which could divert resources to those who could read and understand their medical notes (or who had the confidence to ask questions) might lead to disparities. Awareness of this, and establishing and testing ways to mitigate this risk would be an important element to consider when introducing shared medical records.
Impact on medical practice
Medical records are not only a patient narrative – of their presentation, their investigations and their progress - but a working medical document which reflects dynamic thinking, [ 53 ] consultations, and acts as a tool for handover and for training [ 54 ]. If doctors do not reflect concerns clearly in the notes for fear of worrying the patient, handover could be compromised, impacting negatively on the patient’s care and training of future doctors.
Strengths and limitations
This review synthesised a wide range of papers from medical, nursing and ethical literatures, and was rigorously conducted. However, it identified only papers written in western cultures, and in English, and the conclusions made here should not be extrapolated to other environments. In addition, 7/10 of the studies were carried out in the USA, where the patient doctor relationship also includes a transactional component – doctors need to ensure that patients know what they are paying for. In other health systems represented in these studies (Canada, Norway, Israel) this is not the case, and so the motivations and repercussions of information sharing may be different.
Conclusions and future directions
These studies - and the timing of their publication - reveal that there is significant growth in the approach of sharing more medical information with patients, and significant variation in the type and quantity of information which is being shared. Empirical work with integrated ethical analysis is needed examining the impact of sharing medical records on patient-doctor and multi-disciplinary team communication, on patient trust, on physician training and on resources. The overarching question is what changes will occur to the role of doctor and patient as a result of routinely sharing more information, and, normatively, if there is a “right” amount of information to share with patients in the hospital setting.
Sharing information is a critical part of clinical practice; changing how it is done could have significant empirical and ethical impacts. This review has highlighted what those potential impacts might be. We recommend that careful evaluation of what is recorded and what care is given – both at individual and societal levels – need to be conducted when changes are made to how information is shared.
Availability of data and materials
There are no further data to present other than that which is presented here.
Abbreviations
Cumulative index of nursing and allied health literature
Elton B Stephens Company (A database search database)
Medical subject headings
United Kingdom
United States of America
Access to Health Records Act 1990, available at https://www.legislation.gov.uk/ukpga/1990/23/contents . Accessed 22 Feb 2020.
House of Commons Library. Legislation and guidance relating to medical records explained by House of Commons Library. NHS Confederation 2015, available at https://www.nhsconfed.org/resources/2015/10/legislation-and-guidance-relating-to-medical-records-explained-by-house-of-commonslibrary%20 . Accessed 22 Feb 2020.
Elbourne D, Richardson M, Chalmers I, Waterhouse I, Holt E. The Newbury maternity care study: a randomized controlled trial to assess a policy of women holding their own obstetric records. Br J Obstet Gynaecol. 1987;94(7):612–9.
Article Google Scholar
Liaw T, Lawrence M, Rendell J. The effect of a computer-generated patient-held medical record summary and/or a written personal health record on patients’ attitudes, knowledge and behaviour concerning health promotion. Fam Pract. 1996;13(3):289–93.
Walker J, Leveille SG, Ngo L, Vodicka E, Darer JD, Dhanireddy S, et al. Inviting patients to read their doctors’ notes: patients and doctors look ahead: patient and physician surveys. Ann Intern Med. 2011;155(12):811–9.
Dijkstra RF, Braspenning JC, Huijsmans Z, Akkermans RP, van Ballegooie E, ten Have P, et al. Introduction of diabetes passports involving both patients and professionals to improve hospital outpatient diabetes care. Diabetes Res Clin Pract. 2005;68(2):126–34.
Ayana M, Pound P, Lampe F, Ebrahim S. Improving stroke patients’ care: a patient held record is not enough. BMC Health Serv Res. 2001;1:1.
Volk RJ, Cass AR, Spann SJ. A randomized controlled trial of shared decision making for prostate cancer screening. Arch Fam Med. 1999;8(4):333–40.
Katz SJ, Lantz PM, Janz NK, Fagerlin A, Schwartz K, Liu L, et al. Patient involvement in surgery treatment decisions for breast cancer. J Clin Oncol. 2005;23(24):5526–33.
Ross SE, Lin CT. The effects of promoting patient access to medical records: a review. J Am Med Inform Assoc. 2003;10(2):129–38.
Ferreira A, Correia A, Silva A, Corte A, Pinto A, Saavedra A, et al. Why facilitate patient access to medical records. Stud Health Technol Inform. 2007;127:77–90.
Google Scholar
Goldzweig CL, Orshansky G, Paige NM, Towfigh AA, Haggstrom DA, Miake-Lye I, et al. Electronic patient portals: evidence on health outcomes, satisfaction, efficiency, and attitudes: a systematic review. Ann Intern Med. 2013;159(10):677–87.
Kelly MM, Coller RJ, Hoonakker PL. Inpatient portals for hospitalized patients and caregivers: a systematic review. J Hosp Med. 2017;20:20.
Prey JE, Woollen J, Wilcox L, Sackeim AD, Hripcsak G, Bakken S, et al. Patient engagement in the inpatient setting: a systematic review. J Am Med Inform Assoc. 2014;21(4):742–50.
Vermeir P, Degroote S, Vandijck D, Van Tiggelen H, Peleman R, Verhaeghe R, et al. The patient perspective on the effects of medical record accessibility: a systematic review. Acta Clin Belg. 2017;72(3):186–94.
Dixon-Woods M, Cavers D, Agarwal S, Annandale E, Arthur A, Harvey J, et al. Conducting a critical interpretive synthesis of the literature on access to healthcare by vulnerable groups. BMC Med Res Methodol. 2006;6:35.
Weinert C. Giving Doctors’ daily Progress notes to hospitalized patients and families to improve patient experience. Am J Med Qual. 2017;32(1):58–65.
Urowitz S, Wiljer D, Apatu E, Eysenbach G, Delenardo C, Harth T, et al. Is Canada ready for patient accessible electronic health records? A national scan. BMC Medical Informatics & Decision Making. 2008;8:33.
Weiss M. For doctors’ eyes only: medical records in two Israeli hospitals. Culture Med Psychiat. 1997;21(3):283–302.
Wibe T, Ekstedt M, Helleso R, Slaughter L. Why do people want a paper copy of their electronic patient record? Stud Health Technol Inform. 2010;160(Pt 1):676–80.
Wibe T, Helleso R, Slaughter L, Ekstedt M. Lay people's experiences with reading their medical record. Soc Sci Med. 2011;72(9):1570–3.
Dykes PC, Carroll DL, Hurley AC, Benoit A, Chang F, Pozzar R, et al. Building and testing a patient-centric electronic bedside communication center. J Gerontol Nurs. 2013;39(1):15–9.
Wilcox LG, Gatewood J, Morris D, Tan DS, Feiner S, Horvitz E. Physician attitudes about patient-facing information displays at an urban emergency department. AMIA Annu Symp Proc. 2010;2010:887–91.
Wilcox L, Morris D, Tan D, Gatewood J. Designing patient-centric information displays for hospitals. Proc SIGCHI Conf Hum Factor Comput Syst. 2010;2010:2123–32.
Vawdrey DK, Wilcox LG, Collins SA, Bakken S, Feiner S, Boyer A, et al. A tablet computer application for patients to participate in their hospital care. AMIA Annu Symp Proc. 2011;2011:1428–35.
Collins SA, Rozenblum R, Leung WY, Morrison CR, Stade DL, McNally K, et al. Acute care patient portals: a qualitative study of stakeholder perspectives on current practices. J Am Med Inform Assoc. 2017;24(e1):e9–e17.
Lee EH, Patel JP, Fortin AHV. Patient-centric medical notes: identifying areas for improvement in the age of open medical records. Patient Educ Couns. 2017;100(8):1608–11.
Grossman LV, Choi SW, Collins S, Dykes PC, O'Leary KJ, Rizer M, et al. Implementation of acute care patient portals: recommendations on utility and use from six early adopters. J Am Med Inform Assoc. 2017;04:04.
O'Leary KJ, Lohman ME, Culver E, Killarney A, Randy Smith G Jr, Liebovitz DM. The effect of tablet computers with a mobile patient portal application on hospitalized patients’ knowledge and activation. J Am Med Inform Assoc. 2016;23(1):159–65.
O'Leary KJ, Sharma RK, Killarney A, O'Hara LS, Lohman ME, Culver E, et al. Patients’ and healthcare providers’ perceptions of a mobile portal application for hospitalized patients. BMC Med Inform Decis Mak. 2016;16(1):123.
Beard L, Schein R, Morra D, Wilson K, Keelan J. The challenges in making electronic health records accessible to patients. J Am Med Inform Assoc. 2012;19(1):116–20.
Davis KA, Smith LB. Ethical considerations about EHR-mediated results disclosure and pathology information presented via patient portals. AMA J Ethics. 2016;18(8):826–32.
Lyles CR, Fruchterman J, Youdelman M, Schillinger D. Legal, practical, and ethical considerations for making online patient portals accessible for all. Am J Public Health. 2017;107(10):1608–11.
Gu Y, Orr M, Warren J, Humphrey G, Day K, Tibby S, et al. Why a shared care record is an official medical record. N Z Med J. 2013;126(1384):109–17.
Spriggs M, Arnold MV, Pearce CM, Fry C. Ethical questions must be considered for electronic health records. J Med Ethics. 2012;38(9):535–9.
Gilhooly ML, McGhee SM. Medical records: practicalities and principles of patient possession. J Med Ethics. 1991;17(3):138–43.
Wibe T, Slaughter L. Patients reading their health records - what emotional factors are involved? Studies in Health Technol Inform. 2009;146:174–8.
Dykes PC, Stade D, Chang F, Dalal A, Getty G, Kandala R, et al. Participatory design and development of a patient-centered toolkit to engage hospitalized patients and care partners in their plan of care. AMIA Annu Symp Proc. 2014;2014:486–95.
Baier A. Trust and Antitrust Ethics. 1986;96:231–60.
Zaner RM. The phenomenon of trust in the patient-physician relationship. In: Pellegrino V, editor. Ethics, trust and the professionals: philosophical and cultural Apects. Langan: Georgetown University Press; 1991.
Lee YY, Lin JL. Trust but verify: the interactive effects of trust and autonomy preferences on health outcomes. Health Care Anal. 2009;17(3):244–60.
O'Neill O. Trust with accountability? J Health Serv Res Policy. 2003;8(1):3–4.
O’Neill O. Autonomy and Trust in Bioethics. Cambridge: Cambridge University Press; 2002.
Book Google Scholar
Entwistle VA, Carter SM, Cribb A, McCaffery K. Supporting patient autonomy: the importance of clinician-patient relationships. J Gen Intern Med. 2010;25(7):741–5.
Childress JF. The place of autonomy in bioethics. Hast Cent Rep. 1990;20(1):12–7.
Sandman L, Munthe C. Shared decision-making and patient autonomy. Theor Med Bioeth. 2009;30(4):289–310.
McGuire LC. Remembering what the doctor said: organization and adults’ memory for medical information. Exp Aging Res. 1996;22(4):403–28.
Judson TJ, Detsky AS, Press MJ. Encouraging patients to ask questions: how to overcome “white-coat silence”. JAMA. 2013;309(22):2325–6.
Watt L. “Her life rests on your shoulders”: doing worry as emotion work in the care of children with diabetes. Glob Qual Nurs Res. 2017;4:2333393617743638.
Elwyn G, Barr PJ, Grande SW. Patients recording clinical encounters: a path to empowerment? Assessment by mixed methods. BMJ Open. 2015;5(8):e008566.
Milliat-Guittard L, Charlois AL, Letrilliart L, Favrel V, Galand-Desme S, Schott AM, et al. Shared medical information: expectations of breast cancer patients. Gynecol Oncol. 2007;107(3):474–81.
Lorenc T, Petticrew M, Welch V, Tugwell P. What types of interventions generate inequalities? Evidence from systematic reviews. J Epidemiol Community Health. 2013;67(2):190–3.
Mamykina L, Vawdrey DK, Stetson PD, Zheng K, Hripcsak G. Clinical documentation: composition or synthesis? J Am Med Inform Assoc. 2012;19(6):1025–31.
Rowlands S, Coverdale S, Callen J. Documentation of clinical care in hospital patients’ medical records: a qualitative study of medical students’ perspectives on clinical documentation education. Health Inf Manag. 2016;45(3):99–106.
Download references
Acknowledgments
We would like to thank Anne-Marie Slowther for helpful conversations which guided us during this research.
Zoe Fritz is funded by the Wellcome trust Grant reference numbers: 208213/Z/17/Z, and WT100577MA. The funding body had no role in the design of the study, collection, analysis, interpretation of data, or of the writing the manuscript.
Author information
Authors and affiliations.
Gonville and Caius College, Cambridge University, Trinity Street, Cambridge, CB2 1TA, UK
Stephanie N. D’Costa
THIS Institute (The Healthcare Improvement Studies Institute), Cambridge University, Clifford Allbutt Building, Cambridge, CB2 0AH, UK
Isla L. Kuhn & Zoë Fritz
You can also search for this author in PubMed Google Scholar
Contributions
The Authors have contributed in the following way: ZF conceived of the study, and designed the research questions. SND and IK constructed the literature search; IK refined it and preformed deduplications. SND and ZF screened all papers and identified those for inclusion. ZF and SND drafted different parts of the manuscript. All three authors reviewed, edited and approved the final manuscript.
Corresponding author
Correspondence to Zoë Fritz .
Ethics declarations
Ethics approval and consent to participate.
Not Applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1..
Full search strategies for both literature searches.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
D’Costa, S.N., Kuhn, I.L. & Fritz, Z. A systematic review of patient access to medical records in the acute setting: practicalities, perspectives and ethical consequences. BMC Med Ethics 21 , 18 (2020). https://doi.org/10.1186/s12910-020-0459-6
Download citation
Received : 28 March 2019
Accepted : 17 February 2020
Published : 02 March 2020
DOI : https://doi.org/10.1186/s12910-020-0459-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
BMC Medical Ethics
ISSN: 1472-6939
- General enquiries: [email protected]
SIGNIFICANCE AND CHALLENGES OF MEDICAL RECORDS: A SYSTEMATIC LITERATURE REVIEW

- Skyline University Nigeria
Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations
- BMC HEALTH SERV RES

- Vistolina Nuuyoma
- Hans Justus Amukugo

- Sufyan Ahmed

- BMC MED INFORM DECIS
- Katrin Klug

- Hanson Levi Usende

- Ameenah Muhammad Ndana

- Nicola T Shaw
- Chris Watts

- Kitty McClanahan
- Marcia J. Weiss
- Record Manag J
- LORRAINE NICHOLSON
- Bernadette Szajna
- INT J MED INFORM

- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
IEEE Account
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.

- This Month in Our History
- Swiftly & Safely
- Mayo Clinic in Arizona: 30th Anniversary
- Mayo Clinic in Florida: 30th Anniversary
- Heritage Books
- Heritage Films
- Upcoming Events
- Places To Visit At Mayo Clinic
- Other Historic Locations
- Falcon Program
- Grab-and-Go History Presentation
- Frequently Asked Questions
- Mayo Clinic Flower of Hope
- Contributions to Medicine
- Mayo Clinic Model of Care
- Mayo Clinic Anesthesiology History
From Paper to Digital: The Medical Record at Mayo Clinic
Length: 7:42
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- J Maxillofac Oral Surg
- v.10(3); 2011 Sep

Management of Medical Records: Facts and Figures for Surgeons
1 Department of Oral and Maxillofacial Surgery, M.M. College of Dental Sciences & Research, M.M. University, Mullana, Ambala, Haryana India
Deepika Bali
2 Department of Periodontics, DAV (C) Dental College, Yamuna Nagar, Haryana India
Nageshwar Iyer
Meenakshi iyer.
3 Department of Periodontics, M.M. College of Dental Sciences and Research, M.M. University, Mullana, Ambala, Haryana India
Medical records are the document that explains all detail about the patient’s history, clinical findings, diagnostic test results, pre and postoperative care, patient’s progress and medication. If written correctly, notes will support the doctor about the correctness of treatment. Inspite of knowing the importance of proper record keeping in India, it is still in the initial stages. Medical records are the one of the most important aspect on which practically almost every medico-legal battle is won or lost. This article discusses the various aspect of record maintenance.
Introduction
A good medical record serves the interest of the medical practitioner as well as his patients. It is very important for the treating doctor to properly document the management of the patient under his care. Medical record keeping has evolved into a science. The key to dispensability of most of the medical negligence claim rest with the quality of the medical records. Record maintenance is the only way for the doctor to prove that the treatment was carried out properly. Medical records are often the only source of the truth. They are likely to be far more reliable than memory.
The management and preservation of the hospital records in Indian context present a very gloomy picture. Despite the intensive effort at national and international level, the fundamental health care needs of the population of the developing countries are still unmet. The lack of basic health data renders difficulties in formulating and applying a rational for the allocation of limited resources that are available for patient care and disease prevention.
It is recommended that more efforts should be made by the institutions/hospital managements, all clinicians and medical record officer to improve the standard of maintenance and preservation of medical records. In this article, we are discussing the various aspects of the medical record management.
Objectives of Maintaining Medical Records
- Monitoring of the actual patient
- Medical research
- Medical/dental or paramedical education
- For insurance cases, personal injury suits, workmen’s compensation case, criminal cases, and will cases
- For malpractice suits
- For medical audit and statistical studies
Altering Medical Records
- While writing the medical notes, as far as possible do not overwrite. If the change is needed, strike the whole sentence. Do not leave ambiguity. Make a habit of signing if change is made. Preferably put the date and time below the signature. Attempting to obliterate the erroneous entry by applying the whitener or scratching through the entry in such a way that the person cannot determine what was written originally written raises the suspicion of someone looking for negligent or inappropriate care [ 1 ].
- Do not alter the notes retrospectively. If something written was inaccurate, misleading or incomplete then insert an additional note as a correction [ 2 ].
- Entries in a medical record should be made on every line. Skipping lines leave the room for tampering with the records [ 1 ].
- Amend on electric record by striking through rather than deleting and overwriting the original entry. After inserting the new note, add date, time and doctor name [ 3 ].
- Correction of the personal identification data of the patient like name, age, father/husband name, and address should only be made on the basis of affidavit attested by notary or 1st class magistrate [ 3 ].
Who has Access to Medical Records?
- Medical records are the property of the hospital or patient’s medical practitioner. It is a confidential communication of the patient and cannot be released without his permission [ 1 ].
- All patients have right to access their records and obtain copy of those records [ 1 ].
- Patient’s legal representative has the right to those records as long as patient has signed a release of records to accompany any request from the legal representative [ 4 ].
- Other health care providers have the right to the records of the patient, if they are directly involved in the care and treatment of the patient [ 4 ].
- Parents of a minor also have access to patient’s medical records [ 4 ].
- Medical records are usually summoned in a court of law in certain cases like-road traffic accident, medical negligence, insurance claim etc. [ 2 ].
- The impersonal documents have been used for research purposes as the identity of the patient is not revealed. Though the identity is not revealed, the research team is privy to patient records and a cause of concern about the confidentiality of the information. Recently a need has been felt to regulate the need of medical research, effectively restricting the manner in which this type of research is conducting. An ethical review is required for using the patient’s data [ 3 ].
Release of Records
- Request for medical records by patient or authorized attendant should be acknowledged and documents should be issued within 72 h [ 3 ].
- Maintain the register of certificates with the detail of medical records issued with at least one identification mark of the patient and his signature [ 5 ].
- Effort should be made to computerize the records for quick retrieval [ 2 ].
- Certain document must be given to the patient as a matter of right. Discharge summary, referral notes, or death summary are important document for the patient. Therefore, these documents must be given without any charge for all including patients who discharge themselves against medical advice [ 3 ].
- Doctors are not under any obligation to produce or surrender their medical records to the police in the absence of valid court warrant [ 6 ].
- A subpoena to produce clinical records is a form of court order. Failure to comply is in contempt of court and may be punished. Medical records which are subpoenaed are to be made over to the court and not to the solicitor who sought the subpoena [ 6 ].
Care while Issuing certain Medical Records
Prescription.
The prescription should be preferably on the OPD slip of the institution or on the letter pad of the doctor. Drug company or chemist prescription pad should never be used. Prescription must contain—patient’s name, age, sex, address and institution/hospital name. Prescribed drug should be preferably in capital letter or else clearly visible. One should mention its strength (especially in paediatric age group), its dose frequency, duration in days, and total quantity (number of tablets and capsules). Below the main drug, also mention other instructions of precautions and what to avoid. If any investigation is advised, do not forget to mention it on the prescription slip and call the patient after the investigation. If patient fails to keep follow up date and if then some complication occurs, then patient is also considered negligent (contributory negligence) [ 1 ].
All reports i.e. lab investigation, X-ray reports, ultrasound reports, computed tomography (CT-scan)/magnetic imaging resonance (MRI) reports, and histo-pathological reports should be issued by a qualified person. Biopsy report should preferably be issued in duplicate so that the referring doctor/hospital can keep the original copy. If the pathologist does not give a duplicate copy the referring doctor should get it xeroxed and should be handed over to the patient.
Referral Notes
Always keep the carbon copy of referral note especially in case of critically ill patient. Referral note should mention the date and time of writing the note. Also write the treatment given.
Discharge Card
Consultant in-charge should himself fill or supervise the discharge card. Condition of the patient on the admission, investigation done, the treatment given and detail advice on discharge should be written on discharge card. Operation notes if mentioned have to be correct otherwise just mention the name of the operation and give separate note in detail if asked for. If any complication is expected after discharge ask the patient to report immediately. Instructions while discharge must be very clear and elaborative. Keep in mind that abbreviations may not be understood by others. Also do not use code messages, sarcasm or poor opinion to the patient.
Certificates
A medical certificate is defined as a document of written evidence vouching for the truth of a fact as determined by the doctor issuing such a document. If medical certificate is admitted in a court of law as evidence and is proved to be false, the issuing doctor is liable for punishment. While issuing a medical certificate following things should be kept in mind,
- Medical certificate should be on institution/doctor letter pad.
- Date, time, and place should be mentioned.
- Issue it only for legitimate purpose and only when necessary.
- It has to be true and clear without any ambiguity.
- There should be an identification mark of the patient, preferably a thumb impression.
- Period of illness should be clearly mentioned.
- Diagnosis disclosure of the diagnosis should be only after the patient’s express consent, unless required by the law
- Doctor should maintain the duplicate copy of every certificate.
How Long to Maintain the Records
- Ideally records of adult patient are maintained for 3 year.
- 21 year for neonatal patient (3 + 18 year).
- For children 18 year of age + 3 year.
- For mentally retarded patient forever till hospital/institution is working.
- From income tax point of view for 7 years.
How to Destroy the Records
- Public notice of destroying the records in English news paper and in one vernacular paper mentioning the specific date up to which destruction will be sought [ 1 ].
- Give a time limit of 1 month for taking away records for those who want the records with written consent [ 1 ].
- Where litigation is going on.
- Where future trouble is expected.
- Mentally ill or retarded patient.
- Pre-litigation process of notice exchange is going on.
Hard Copy Only
Computers are now widely used in institution/hospitals for electronic patient records but still hard copy is required for following documents [ 1 ]
- Consent need to be on hard copy.
- Referral to doctor need hard copy.
- Police case need hard copy.
- Certificate of fitness should be on hard copy.
Problem of Record Management
There are many problems faced by institution/hospital for the proper maintenance of the records. 1. Constant revision of the outdated form is needed [ 2 ]. 2. Always trained personnel are needed for the maintenance [ 2 ]. 3. Inactive records need storage at appropriate place [ 7 ]. 4. There must be a need of determination of record retention [ 7 ]. 5. Unwanted records must be destroyed [ 8 ]. 6. Record storage entail into 2 stages. A. Moving the records from active to inactive file and from there to storage room. B. Destruction and disposal of the unimportant records [ 8 ].
There are various type of damage which may be found in paper documentation like-aged paper may become weak, colour alteration from white to yellow, dirt and dust may be present on the surface, insect and fungus is a big threat for the records, if paper is kept folded, it may become weak at the crease, dampness and water leakage in storage room also destroy the paper.
Proper Preservation of the Medical Records
Collect all the records and classify them according to the different section [ 7 ]. Protect the records from insect attack. Spray insecticide or place naphthalene balls over shelves to preserve the records. Plan a periodical checking for the records [ 3 ]. Proper care should be observed while handling the records. Fire extinguisher should be available in record room. Protect all records from dampness, water, and from hot and dry climate [ 8 ]. Records should be kept in dust free area. Windows and ventilators should be properly covered with frames as safeguard against sabotage. Destroy the records as per the regulation established for retention of records.
Medical records form an important part of a patient management. It is important for the doctor and medical establishment to properly maintain the records of the patient for 2 important reasons. First one is that it helps in proper evaluation of the patient and to plan treatment protocol. Second is that the legal system relies mainly on documentary evidence in cases of medical negligence. Therefore, medical records should be properly written and preserved to serve the interest of doctor as well as his patient.

IMAGES
VIDEO
COMMENTS
Effects of Electronic Health Record Implementation and ...
Abstract. Electronic Health Records (EHRs) have revolutionized healthcare delivery, offering numerous benefits for patient care and outcomes. This comprehensive review examines the impact of EHRs ...
The Use of Medical Records in Research: What Do ...
Electronic Health Records: Then, Now, and in the Future
Security and privacy of electronic health records: Concerns ...
The misuse of medical records is not only revealed on paper-based medical records but also occurs in electronic medical record. According to the research from (Harande, 2018), the medical records ...
A 2019 poll by the Henry J. Kaiser Family Foundation, a non-profit health-care advocacy organization in San Francisco, California, found that 45% of US citizens think that electronic health ...
An electronic health record (EHR) is the systematized collection of patient and population electronically stored health information in a digital format 1 and providing patients with access to EHRs has the potential to decrease these costs, improve self-care, quality of care, and health and patient-centered outcome. 1, 2.
papers between databases, where we found n o connection to the subject of concern, just th e medical part, we removed articles through a series of iterations of filterin g and sorting. Reports ...
A large language model for electronic health records
1. It reduces time spent on paper documentation. 2. Massive data can be gathered from the EHR, which can be very useful for research and analysis. It takes time to get used to the EHR system and requires plenty of documentation, making EHR less efficient. Nurse practitioners: 1. EHR promotes an understanding of the doctor's plan in real-time. 2.
The proliferation of electronic health records (EHRs) spurred on by federal government incentives over the past few decades has resulted in greater than an 80% adoption-rate at hospitals [] and close to 90% in office-based practices [] in the United States.A natural consequence of the availability of electronic health data is the conduct of research with these data, both observational and ...
Electronic medical records - The good, the bad and the ugly
A systematic review of patient access to medical records in the ...
Electronic health records (EHRs) are a significant advancement over paper records. However, the full potential of EHRs for improving care quality, patient outcomes, surveillance, and research in cancer care is yet to be realized. The organic evolution of EHRs has resulted in a number of unanticipated consequences including increased time spent by clinicians interfacing with the EHR for daily ...
Furthermore, this system allows for easy location and access for healthcare personnel. 3,4 On the other hand, paper medical records can be unsustainable, highly inefficient, time-consuming, and space ... 74 studies were conducted in developed economies, and 58 non-empirical research papers were excluded. The authors then reviewed the remaining ...
Value of the Electronic Medical Record for Hospital Care
Conceived as a solution to problems inherent in paper records, the development of the electronic health record and electronic medical record has been rapid compared with that of the modern medical record itself, as described extensively elsewhere. 1-3,23-28 Stimulated in Europe by national health insurance systems and in the US by demands of ...
Abstract. Medical records are a vi tal asset in ensuring that hospitals are run effectivel y and. efficiently. They support clinical dec ision-making, provide evidence o f policies and. support ...
The medical records are the records of the patient's disease occurrence, development, examination, diagnosis, and treatment. Medical records are an important part of medical care, teaching, prevention, research and development. They are the basic support conditions for hospital management. It is difficult to keep the traditional paper medical records. It is also difficult to query the paper ...
Medical Records: A Historical Narrative - PMC
Learn how Mayo Clinic's medical record evolved from paper ledgers to the electronic tool of today.
Medical records are the document that explains all detail about the patient's history, clinical findings, diagnostic test results, pre and postoperative care, patient's progress and medication. If written correctly, notes will support the doctor about the correctness of treatment. Inspite of knowing the importance of proper record keeping ...